1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
1OAS
 
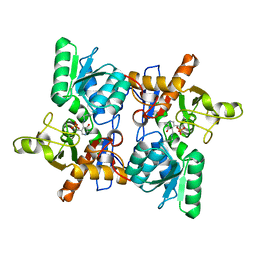 | | O-ACETYLSERINE SULFHYDRYLASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Rao, G.S.J, Hohenester, E, Schnackerz, K.D, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-01-29 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 283, 1998
|
|
3SS9
 
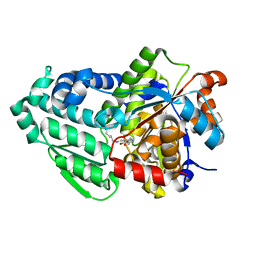 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
3SS7
 
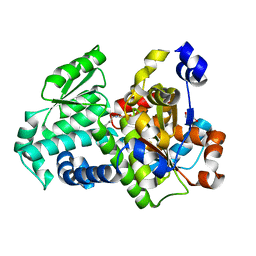 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
1H7X
 
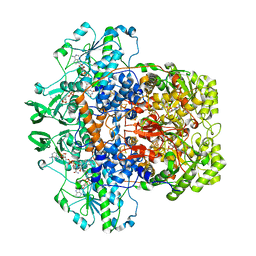 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
1H7W
 
 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GT8
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1D6S
 
 | | CRYSTAL STRUCTURE OF THE K41A MUTANT OF O-ACETYLSERINE SULFHYDRYLASE COMPLEXED IN EXTERNAL ALDIMINE LINKAGE WITH METHIONINE | | Descriptor: | METHIONINE, O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Tai, C.H, Ristroph, C.M, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-10-15 | | Release date: | 2000-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 291, 1999
|
|
1FCJ
 
 | | CRYSTAL STRUCTURE OF OASS COMPLEXED WITH CHLORIDE AND SULFATE | | Descriptor: | CHLORIDE ION, O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Burkhard, P, Tai, C, Jansonius, J.N, Cook, P.F. | | Deposit date: | 2000-07-18 | | Release date: | 2000-10-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: structure of the enzyme with chloride bound.
J.Mol.Biol., 303, 2000
|
|
