1IVY
 
 | | PHYSIOLOGICAL DIMER HPP PRECURSOR | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudenko, G, Bonten, E, D'Azzo, A, Hol, W.G.J. | | Deposit date: | 1996-06-12 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of the human 'protective protein': structure of the precursor form suggests a complex activation mechanism.
Structure, 3, 1995
|
|
1N7D
 
 | | Extracellular domain of the LDL receptor | | Descriptor: | 12-TUNGSTOPHOSPHATE, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Rudenko, G, Henry, L, Henderson, K, Ichtchenko, K, Brown, M.S, Goldstein, J.L, Deisenhofer, J. | | Deposit date: | 2002-11-13 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the LDL receptor extracellular domain at endosomal pH
Science, 298, 2002
|
|
1C4R
 
 | | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | | Descriptor: | NEUREXIN-I BETA | | Authors: | Rudenko, G, Nguyen, T, Chelliah, Y, Sudhof, T.C, Deisenhofer, J. | | Deposit date: | 1999-09-28 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Cell(Cambridge,Mass.), 99, 1999
|
|
2R16
 
 | |
3QCW
 
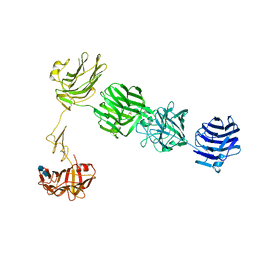 | | Structure of neurexin 1 alpha (domains LNS1-LNS6), no splice inserts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neurexin-1-alpha | | Authors: | Rudenko, G. | | Deposit date: | 2011-01-17 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of Neurexin 1 alpha Reveals Features Promoting a Role as Synaptic Organizer
Structure, 19, 2011
|
|
2R1D
 
 | |
2R1B
 
 | |
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7UCC
 
 | | Transcription factor FosB/JunD bZIP domain in the reduced form | | Descriptor: | CHLORIDE ION, ETHANOL, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
6CW1
 
 | |
2H0B
 
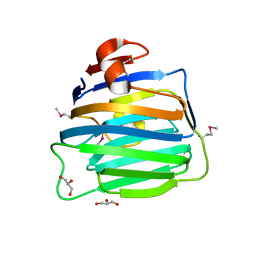 | | Crystal Structure of the second LNS/LG domain from Neurexin 1 alpha | | Descriptor: | CALCIUM ION, GLYCEROL, Neurexin-1-alpha | | Authors: | Sheckler, L.R, Henry, L, Sugita, S, Sudhof, T.C, Rudenko, G. | | Deposit date: | 2006-05-14 | | Release date: | 2006-06-20 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Second LNS/LG Domain from Neurexin 1{alpha}: Ca2+ binding and the effects of alternative splicing
J.Biol.Chem., 281, 2006
|
|
7KBU
 
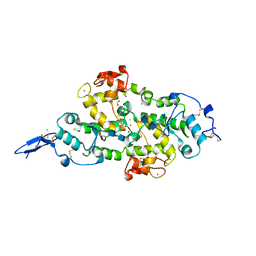 | | Structure of Hevin FS-EC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Machius, M, Fan, S, Rudenko, G. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Interplay between hevin, SPARC, and MDGAs: Modulators of neurexin-neuroligin transsynaptic bridges.
Structure, 29, 2021
|
|
6UCM
 
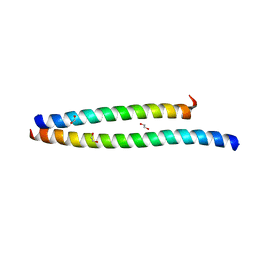 | | Transcription factor DeltaFosB bZIP domain self-assembly, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCL
 
 | |
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
3R05
 
 | |
6U6T
 
 | | Neuronal growth regulator 1 (NEGR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuronal growth regulator 1, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
5V5W
 
 | |
5VPC
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-II crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M.C, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPA
 
 | | Transcription factor FosB/JunD bZIP domain | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPF
 
 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
