4X4J
 
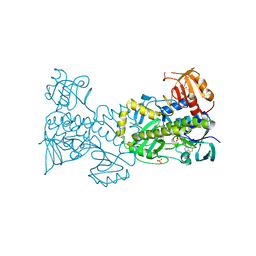 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
3FMW
 
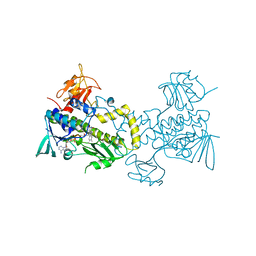 | | The crystal structure of MtmOIV, a Baeyer-Villiger monooxygenase from the mithramycin biosynthetic pathway in Streptomyces argillaceus. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Beam, M.P, Wang, C, Rohr, J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway .
Biochemistry, 48, 2009
|
|
5JVW
 
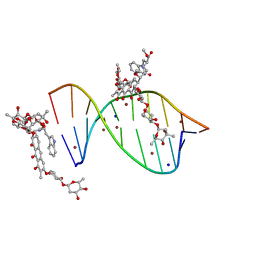 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
3POP
 
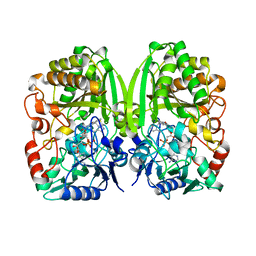 | | The crystal structure of GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GilR oxidase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-23 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
3PQB
 
 | | The crystal structure of pregilvocarcin in complex with GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | (1R)-1,4-anhydro-6-deoxy-1-[(6R)-8-ethenyl-1,6-dihydroxy-10,12-dimethoxy-6H-dibenzo[c,h]chromen-4-yl]-D-galactitol, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
6OVQ
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | Descriptor: | GLYCEROL, Putative Side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OW0
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6VGG
 
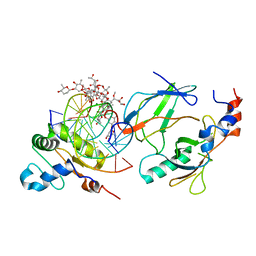 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2020-01-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
5JW0
 
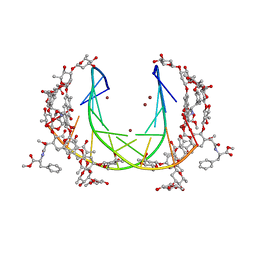 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGTACCCT | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*TP*AP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
5JW2
 
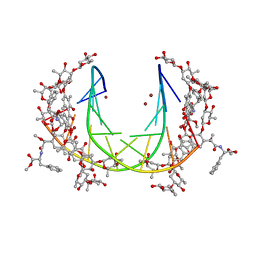 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGATCCCT | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*AP*TP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
4K5R
 
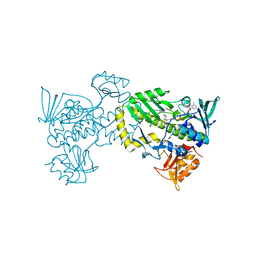 | | The 2.0 angstrom crystal structure of MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
4K5S
 
 | | The crystal structure of premithramycin B in complex with MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase, premithramycin B | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
4RVH
 
 | | Crystal structure of MtmC in complex with SAH and TDP-4-keto-D-olivose | | Descriptor: | D-mycarose 3-C-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVG
 
 | | Crystal structure of MtmC in complex with SAM and TDP | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVD
 
 | | Crystal structure of MtmC in complex with SAM | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVF
 
 | | Crystal structure of MtmC in complex with TDP | | Descriptor: | D-mycarose 3-C-methyltransferase, THYMIDINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RV9
 
 | | Crystal structure of MtmC in complex with SAH | | Descriptor: | ACETATE ION, CHLORIDE ION, D-mycarose 3-C-methyltransferase, ... | | Authors: | Hou, C, Chen, J.-M, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2014-11-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
5NV8
 
 | | Structural basis for EarP-mediated arginine glycosylation of translation elongation factor EF-P | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, EF-P arginine 32 rhamnosyl-transferase | | Authors: | Macosek, J, Krafczyk, R, Jagtap, P.K.A, Lassaka, J, Hennig, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P.
MBio, 8, 2017
|
|
4RIE
 
 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIF
 
 | | Landomycin Glycosyltransferase LanGT2, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIG
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac | | Descriptor: | Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIH
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RII
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, TDP complex | | Descriptor: | Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6VGD
 
 | |
