1C95
 
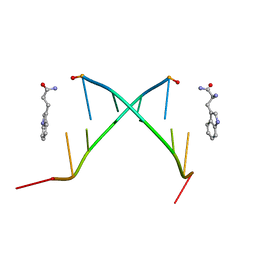 | |
1KSE
 
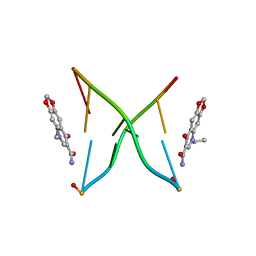 | | Solution Structure of a quinolone-capped DNA duplex | | Descriptor: | 5'-D(*(5AT)P*GP*CP*GP*CP*A)-3', OXOLINIC ACID | | Authors: | Tuma, J, Connors, W.H, Stitelman, D.H, Richert, C. | | Deposit date: | 2002-01-12 | | Release date: | 2002-05-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | On the effect of covalently appended quinolones on termini of DNA duplexes.
J.Am.Chem.Soc., 124, 2002
|
|
1X6W
 
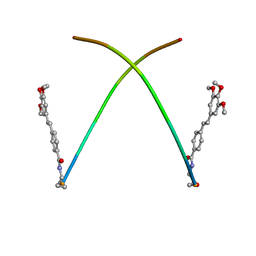 | |
1ON5
 
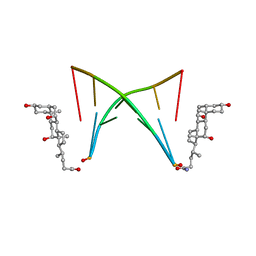 | |
2KK5
 
 | | High Fidelity Base Pairing at the 3'-Terminus | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*(2AU))-3', 9,10-dioxo-9,10-dihydroanthracene-2-carboxamide | | Authors: | Patra, A, Richert, C. | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | High fidelity base pairing at the 3'-terminus.
J.Am.Chem.Soc., 131, 2009
|
|
2BQ2
 
 | | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*2AU)-3', NALIDIXIC ACID | | Authors: | Siegmund, K, Maheshwary, S, Narayanan, S, Connors, W, Richert, M. | | Deposit date: | 2005-04-26 | | Release date: | 2006-08-03 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it.
Nucleic Acids Res., 33, 2005
|
|
6XHC
 
 | | Structure of glycinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]ethanoic acid, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHE
 
 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
