2H85
 
 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FSD
 
 | |
1P5R
 
 | | Formyl-CoA Transferase in complex with Coenzyme A | | Descriptor: | COENZYME A, Formyl-coenzyme A transferase | | Authors: | Ricagno, S, Jonsson, S, Richards, N, Lindqvist, Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Formyl-CoA Transferase encloses the CoA binding site at the interface of an interlocked dimer
Embo J., 22, 2003
|
|
1P5H
 
 | |
2V2K
 
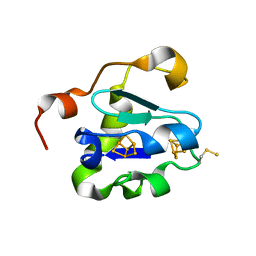 | | THE CRYSTAL STRUCTURE OF FDXA, A 7FE FERREDOXIN FROM MYCOBACTERIUM SMEGMATIS | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FERREDOXIN | | Authors: | Ricagno, S, de Rosa, M, Aliverti, A, Zanetti, G, Bolognesi, M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Fdxa, a 7Fe Ferredoxin from Mycobacterium Smegmatis.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
1R0K
 
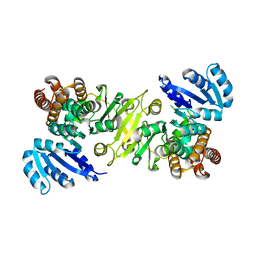 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Zymomonas mobilis | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ACETATE ION | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
1R0L
 
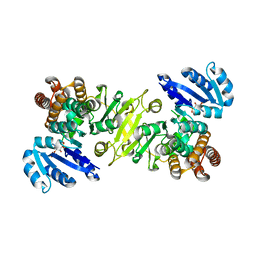 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from zymomonas mobilis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ricagno, S, Grolle, S, Bringer-Meyer, S, Sahm, H, Lindqvist, Y, Schneider, G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution.
Biochim.Biophys.Acta, 1698, 2004
|
|
3DHJ
 
 | | Beta 2 microglobulin mutant W60C | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
3DHM
 
 | | Beta 2 microglobulin mutant D59P | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
3EKC
 
 | |
3F02
 
 | |
3F5N
 
 | |
2Z9T
 
 | | Crystal structure of the human beta-2 microglobulin mutant W60G | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Bolognesi, M, Bellotti, V, Corazza, A, Rennella, E, Gural, D, Mimmi, M.C, Betto, E, Pucillo, C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties
J.Mol.Biol., 378, 2008
|
|
1VGR
 
 | | Formyl-CoA transferase mutant Asp169 to Glu | | Descriptor: | COENZYME A, Formyl-coenzyme A transferase | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1VGQ
 
 | | Formyl-CoA transferase mutant Asp169 to Ala | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
6TCC
 
 | | Crystal structure of Salmo salar RidA-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
6TCD
 
 | | Crystal structure of Salmo salar RidA-2 | | Descriptor: | ACETATE ION, Ribonuclease UK114, SULFATE ION | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
3QDA
 
 | | Crystal structure of W95L beta-2 microglobulin | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Ricagno, S, Bellotti, V, Bolognesi, M. | | Deposit date: | 2011-01-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The two tryptophans of beta2-microglobulin have distinct roles in function and folding and might represent two independent responses to evolutionary pressure.
BMC Evol Biol, 11, 2011
|
|
4FXL
 
 | | Crystal structure of the D76N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Bellotti, V, Pepys, M.B, Stoppini, M, Bolognesi, M. | | Deposit date: | 2012-07-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hereditary systemic amyloidosis due to Asp76Asn variant beta-2-microglobulin.
N.Engl.J.Med., 366, 2012
|
|
1T4C
 
 | | Formyl-CoA Transferase in complex with Oxalyl-CoA | | Descriptor: | COENZYME A, Formyl-CoA:oxalate CoA-transferase, OXALIC ACID | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1T3Z
 
 | | Formyl-CoA Tranferase mutant Asp169 to Ser | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
8CPE
 
 | |
8P88
 
 | |
8P89
 
 | |
6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | Descriptor: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | Authors: | Costanzi, E, Sormanni, P, Ricagno, S. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
