6VZD
 
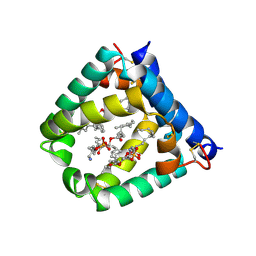 | | N-terminal domain of mouse surfactant protein B (K46E/R51E mutant) with bound lipid | | Descriptor: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | Authors: | Rapoport, T.A, Bodnar, N.O. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
6VZE
 
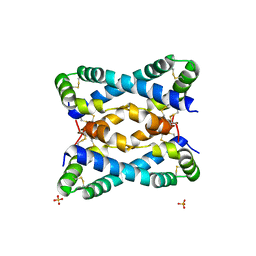 | |
6VYN
 
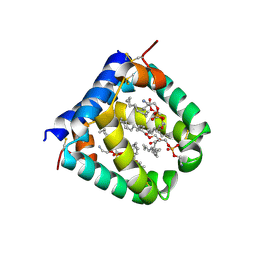 | | N-terminal domain of mouse surfactant protein B with bound lipid, wild type | | Descriptor: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | Authors: | Rapoport, T.A, Bodnar, N.O. | | Deposit date: | 2020-02-27 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
6W1B
 
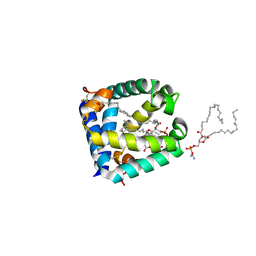 | |
6VZ0
 
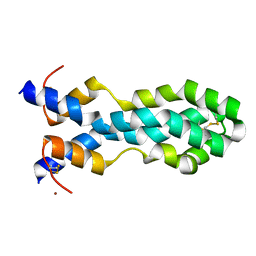 | |
4V4N
 
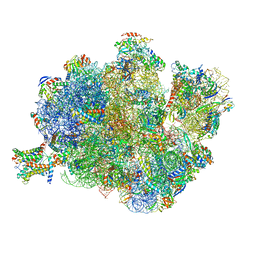 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
6ITC
 
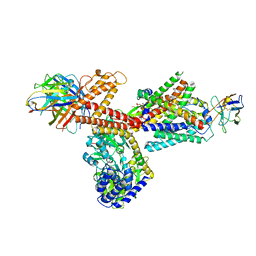 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
7R9D
 
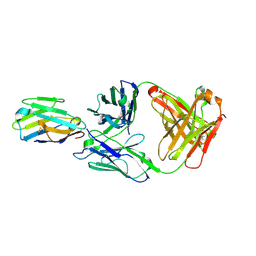 | | Crystal structure of Nb_0 in complex with Fab_8D3 | | Descriptor: | Fab 8D3 heavy chain, Fab 8D3 light chain, Nanobody N0 | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RXD
 
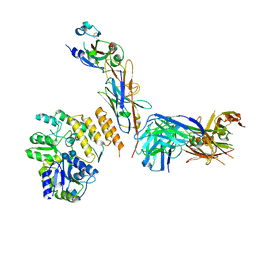 | | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | | Descriptor: | Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RXC
 
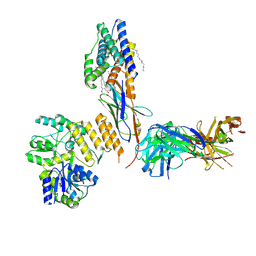 | | CryoEM structure of KDELR with Legobody | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ER lumen protein-retaining receptor 2, Fab_8D3_2 heavy chain, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MBK
 
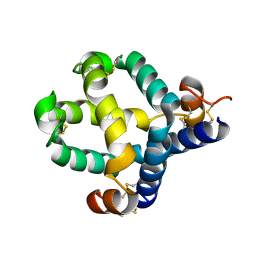 | |
4YS0
 
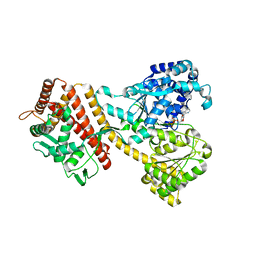 | |
5L0Y
 
 | |
5L0W
 
 | |
6CDD
 
 | |
6CHS
 
 | | Cdc48-Npl4 complex in the presence of ATP-gamma-S | | Descriptor: | MAGNESIUM ION, Npl4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kim, K.H, Bodnar, N.O, Walz, T, Rapoport, T.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2IBM
 
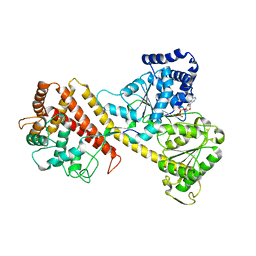 | |
5E9U
 
 | | Crystal structure of GtfA/B complex bound to UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, Glycosyltransferase-stabilizing protein Gtf2, ... | | Authors: | Chen, Y, Rapoport, T.A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E9T
 
 | | Crystal structure of GtfA/B complex | | Descriptor: | Glycosyltransferase Gtf1, Glycosyltransferase-stabilizing protein Gtf2, MAGNESIUM ION | | Authors: | Chen, Y, Rapoport, T.A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Mechanism of a cytosolic O-glycosyltransferase essential for the synthesis of a bacterial adhesion protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EUL
 
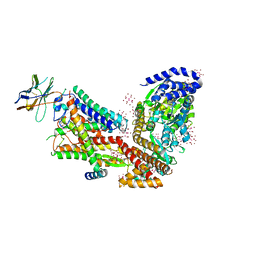 | | Structure of the SecA-SecY complex with a translocating polypeptide substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AYC08, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Li, L, Park, E, Ling, J, Ingram, J, Ploegh, H, Rapoport, T.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of a substrate-engaged SecY protein-translocation channel.
Nature, 531, 2016
|
|
6ND1
 
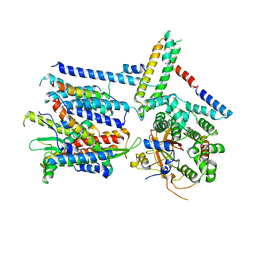 | | CryoEM structure of the Sec Complex from yeast | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Wu, X, Cabanos, C, Rapoport, T.A. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the post-translational protein translocation machinery of the ER membrane.
Nature, 566, 2019
|
|
6OA9
 
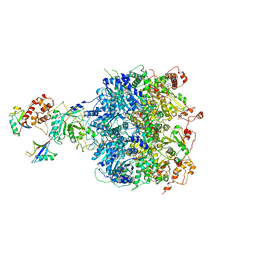 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OAA
 
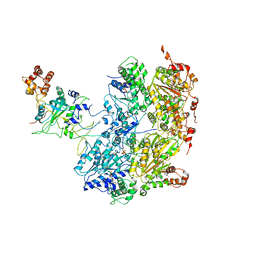 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OAB
 
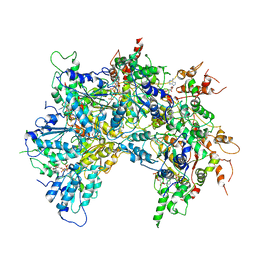 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
4MLS
 
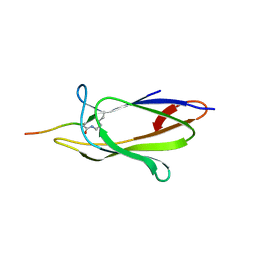 | | Crystal structure of the SpyTag and SpyCatcher-deltaN1 complex | | Descriptor: | Fibronectin binding protein, SpyTag | | Authors: | Li, L, Fierer, J.O, Rapoport, T.A, Howarth, M. | | Deposit date: | 2013-09-06 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag.
J.Mol.Biol., 426, 2014
|
|
