5LF2
 
 | | Crystal structure of laminin beta2 LE5-LF-LE6 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Laminin subunit beta-2 | | Authors: | Pulido, D, Hohenester, E. | | Deposit date: | 2016-06-30 | | Release date: | 2016-07-27 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic analysis of the laminin beta 2 short arm reveals how the LF domain is inserted into a regular array of LE domains.
Matrix Biol., 57-58, 2017
|
|
5MC9
 
 | |
6FZV
 
 | |
6FZW
 
 | |
4X09
 
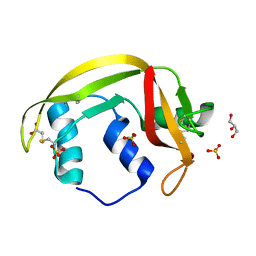 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
4A2O
 
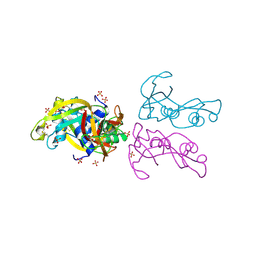 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH SULFATE ANIONS | | Descriptor: | EOSINOPHIL CATIONIC PROTEIN, SULFATE ION | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
4A2Y
 
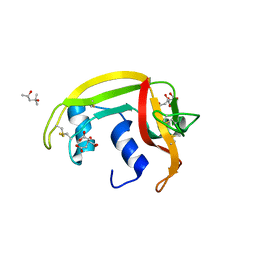 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH CITRATE ANIONS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
7PHW
 
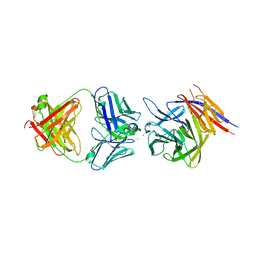 | | PfCyRPA bound to monoclonal antibody Cy.004 Fab fragment | | Descriptor: | CALCIUM ION, Cysteine-rich protective antigen, Monoclonal antibody Cy.004 heavy chain, ... | | Authors: | Ragotte, R.J, Higgins, M.K. | | Deposit date: | 2021-08-18 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Heterotypic interactions drive antibody synergy against a malaria vaccine candidate.
Nat Commun, 13, 2022
|
|
7PHV
 
 | | PfCyRPA bound to monoclonal antibody Cy.007 Fab fragment | | Descriptor: | AMMONIUM ION, Cysteine-rich protective antigen, Monoclonal antibody Cy.007 heavy chain, ... | | Authors: | Ragotte, R.J, Higgins, M.K. | | Deposit date: | 2021-08-18 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Heterotypic interactions drive antibody synergy against a malaria vaccine candidate.
Nat Commun, 13, 2022
|
|
7PI2
 
 | | PfCyRPA bound to monoclonal antibody Cy.003 Fab fragment | | Descriptor: | Cysteine-rich protective antigen, Monoclonal antibody Cy.003 heavy chain, Monoclonal antibody Cy.003 light chain | | Authors: | Ragotte, R.J, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Heterotypic interactions drive antibody synergy against a malaria vaccine candidate.
Nat Commun, 13, 2022
|
|
7PI7
 
 | | PfCyRPA bound to monoclonal antibody Cy.002 Fab fragment | | Descriptor: | Cysteine-rich protective antigen, Monoclonal antibody Cy.002 heavy chain, Monoclonal antibody Cy.002 light chain | | Authors: | Ragotte, R.J, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Heterotypic interactions drive antibody synergy against a malaria vaccine candidate.
Nat Commun, 13, 2022
|
|
7PI3
 
 | |
7PHU
 
 | |
6RCV
 
 | | PfRH5 bound to monoclonal antibodies R5.011 and R5.016 | | Descriptor: | R5.011 heavy chain, R5.011 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCO
 
 | | PfRH5-binding monoclonal antibody R5.004 | | Descriptor: | ACETATE ION, GLYCEROL, R5.004 heavy chain, ... | | Authors: | Alanine, D.G.W, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCU
 
 | | PfRH5 bound to monoclonal antibodies R5.004 and R5.016 | | Descriptor: | R5.004 heavy chain, R5.004 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCQ
 
 | | PfRH5-binding monoclonal antibody R5.011 | | Descriptor: | ACETATE ION, CACODYLATE ION, R5.011 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCS
 
 | | PfRH5-binding monoclonal antibody R5.016 | | Descriptor: | ACETATE ION, R5.016 heavy chain, R5.016 light chain, ... | | Authors: | Alanine, D.W.G, Jamwal, A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
5OAB
 
 | | A novel crystal form of human RNase6 at atomic resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4OXF
 
 | |
4OXB
 
 | |
6ENP
 
 | | Atomic resolution structure of human RNase 6 in the presence of phosphate anions in P21 space group. | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Ribonuclease K6, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.042 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
