2MN2
 
 | | 3D structure of YmoB, a modulator of biofilm formation | | Descriptor: | YmoB | | Authors: | Marimon, O, Cordeiro, T.N, Amata, I, Pons, M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An oxygen-sensitive toxin-antitoxin system.
Nat Commun, 7, 2016
|
|
1KTX
 
 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
2KTX
 
 | | COMPLETE KALIOTOXIN FROM ANDROCTONUS MAURETANICUS MAURETANICUS, NMR, 18 STRUCTURES | | Descriptor: | KALIOTOXIN | | Authors: | Gairi, M, Romi, R, Fernandez, I, Rochat, H, Martin-Eauclaire, M.-F, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | 3D structure of kaliotoxin: is residue 34 a key for channel selectivity?
J.Pept.Sci., 3, 1997
|
|
2LEV
 
 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
2MW2
 
 | | Hha-H-NS46 charge zipper complex | | Descriptor: | DNA-binding protein H-NS, Hemolysin expression-modulating protein Hha | | Authors: | Cordeiro, T.N, Garcia, J, Bernado, P, Millet, O, Pons, M. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-29 | | Last modified: | 2018-01-10 | | Method: | SOLUTION NMR | | Cite: | A Three-protein Charge Zipper Stabilizes a Complex Modulating Bacterial Gene Silencing.
J. Biol. Chem., 290, 2015
|
|
6HC3
 
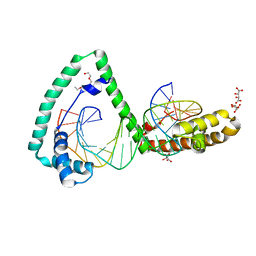 | | TFAM bound to Site-X | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*GP*TP*GP*GP*AP*AP*AP*TP*TP*TP*TP*TP*TP*GP*TP*TP*AP*G)-3'), DNA/RNA (5'-D(*TP*AP*AP*CP*AP*AP*AP*AP*AP*AP*TP*TP*TP*CP*CP*AP*CP*CP*AP*AP*AP*C)-3'), L(+)-TARTARIC ACID, ... | | Authors: | Fernandez-Millan, P, Cuppari, A, Tarres-Sole, A, Rubio-Cosials, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
7AIV
 
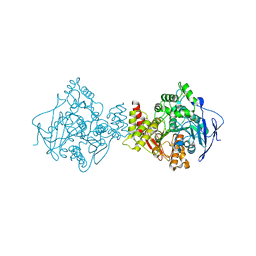 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 4-{[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]methyl}-N-(4-hydroxy-3-methoxybenzyl)benzamide | | Descriptor: | 4-{[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]methyl}-N-(4-hydroxy-3-methoxybenzyl)benzamide, Acetylcholinesterase | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIW
 
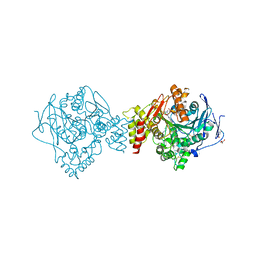 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with (E)-10-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)-6-decenamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIS
 
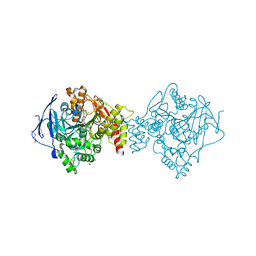 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 6-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)hexanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)hexanamide, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIX
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | Descriptor: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIY
 
 | | Crystal structure of human butyrylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | Descriptor: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Cholinesterase | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIU
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 8-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)octanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)octanamide, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIT
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 7-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)heptanamide | | Descriptor: | 7-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)heptanamide, Acetylcholinesterase | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
6HB4
 
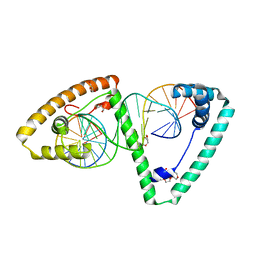 | | TFAM in Complex with Site-Y | | Descriptor: | DNA (5'*CP*TP*GP*TP*GP*CP*AP*GP*AP*CP*AP*TP*TP*CP*AP*AP*TP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*AP*TP*TP*GP*AP*AP*TP*GP*TP*CP*TP*GP*CP*AP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Cuppari, A, Fernandez-Millan, P, Rubio-Cosials, A, Tarres-Sole, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
5DKA
 
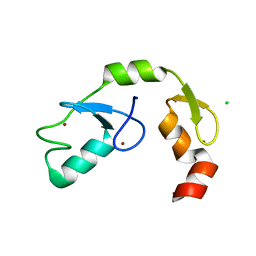 | | A C2HC zinc finger is essential for the activity of the RING ubiquitin ligase RNF125 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF125, MAGNESIUM ION, ... | | Authors: | Boer, D.R, Coll, M, Bijlmakers, M.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125.
Sci Rep, 6, 2016
|
|
5SXR
 
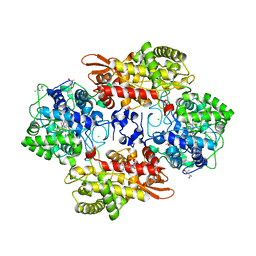 | | Crystal structure of B. pseudomallei KatG with NAD bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases.
J. Biol. Chem., 285, 2010
|
|
5SXX
 
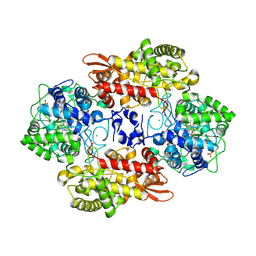 | |
5SXQ
 
 | |
5SXS
 
 | |
5SXW
 
 | |
5SXT
 
 | |
1TXE
 
 | | Solution structure of the active-centre mutant Ile14Ala of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus carnosus | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Moeglich, A, Koch, B, Hengstenberg, W, Brunner, E, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-07-04 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus
Eur.J.Biochem., 271, 2004
|
|
2JZP
 
 | | NMR solution structure of Kx5Q ProtL mutant | | Descriptor: | Protein L | | Authors: | Lopez-Mendez, N, Tadeo, X, Castano, D, Pons, M, Millet Aguilar-Galindo, O. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-13 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Protein stabilization and the hofmeister effect: the role of hydrophobic solvation
Biophys.J., 97, 2009
|
|
2KAC
 
 | |
