4ZDA
 
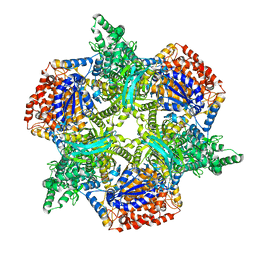 | |
2F82
 
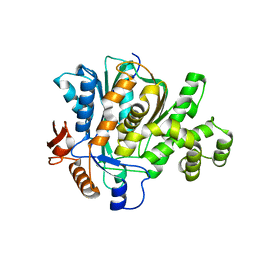 | |
2FA0
 
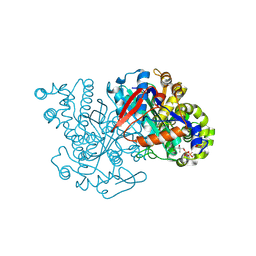 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F9A
 
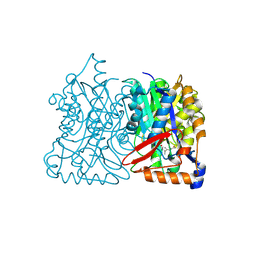 | | HMG-CoA synthase from Brassica juncea in complex with F-244 | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, 3-Hydroxy-3-methylglutaryl coenzyme A synthase 1 | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-05 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FA3
 
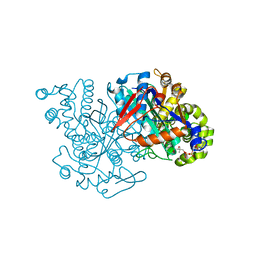 | | HMG-CoA synthase from Brassica juncea in complex with acetyl-CoA and acetyl-cys117. | | Descriptor: | ACETYL COENZYME *A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5MTN
 
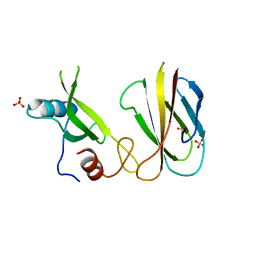 | | Monobody Mb(Lck_1) bound to Lck-Sh2 | | Descriptor: | Monobody Mb(Lck_1), SULFATE ION, Tyrosine-protein kinase Lck | | Authors: | Pojer, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
5MTM
 
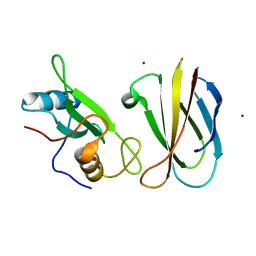 | | Monobody Mb(Lck_3) bound to Lck-SH2 domain | | Descriptor: | Monobody Mb(Lck_3), Tyrosine-protein kinase Lck, ZINC ION | | Authors: | Pojer, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
2Q6K
 
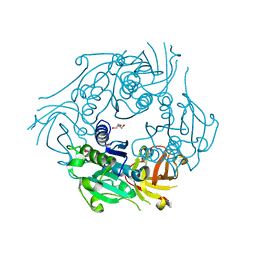 | | SalL with adenosine | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, chlorinase | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6L
 
 | | SalL double mutant Y70T/G131S with CLDA and L-MET | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Hypothetical protein, METHIONINE | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
3RIM
 
 | | Crystal structure of mycobacterium tuberculosis Transketolase (Rv1449c) | | Descriptor: | GLYCEROL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Pojer, F, Fullam, E, Jones, T.A, Cole, S.T. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and function of the transketolase from Mycobacterium tuberculosis and comparison with the human enzyme.
Open Biol, 2, 2012
|
|
4BGE
 
 | |
4BGI
 
 | | Crystal structure of InhA(S94A) mutant in complex with OH-141 | | Descriptor: | 3-hydroxy-N-[(2R,5R,6S,9S,10S,11R)-10-hydroxy-5,11-dimethyl-3,7,12-trioxo-2-(propan-2-yl)-9-(pyridin-3-ylmethyl)-1,4-dioxa-8-azacyclododecan-6-yl]pyridine-2-carboxamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pojer, F, Hartkoorn, R.C, Cole, S.T. | | Deposit date: | 2013-03-27 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridomycin bridges the NADH- and substrate-binding pockets of the enoyl reductase InhA.
Nat. Chem. Biol., 10, 2014
|
|
4DQU
 
 | | Mycobacterium tuberculosis InhA-D148G mutant in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
4DTI
 
 | | Mycobacterium tuberculosis InhA-S94A mutant in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
4DRE
 
 | | Mycobacterium tuberculosis InhA in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pojer, F, Hartkoorn, R.C, Boy, S, Cole, S.T. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Towards a new tuberculosis drug: pyridomycin - nature's isoniazid.
EMBO Mol Med, 4, 2012
|
|
3BWO
 
 | | L-tryptophan aminotransferase | | Descriptor: | L-tryptophan aminotransferase | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for the shade avoidance response of plants
To be Published
|
|
3BWN
 
 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
6HTF
 
 | |
6O02
 
 | |
6O03
 
 | |
6QGS
 
 | | Crystal structure of APT1 bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
8AQU
 
 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQT
 
 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQS
 
 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
