8DDJ
 
 | | Open MscS in PC14.1 Nanodiscs | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Reddy, B.G, Bavi, N, Perozo, E. | | Deposit date: | 2022-06-18 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | State-specific morphological deformations of the lipid bilayer explain mechanosensitive gating of MscS ion channels.
Elife, 12, 2023
|
|
8UC1
 
 | |
1JQ2
 
 | |
1JQ1
 
 | |
1F6G
 
 | | POTASSIUM CHANNEL (KCSA) FULL-LENGTH FOLD | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL | | Authors: | Cortes, D.M, Perozo, E. | | Deposit date: | 2000-06-21 | | Release date: | 2001-02-21 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating.
J.Gen.Physiol., 117, 2001
|
|
1ZWI
 
 | | Structure of mutant KcsA potassium channel | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Cordero-Morales, J.F, Cuello, L.G, Zhao, Y, Jogini, V, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2005-06-03 | | Release date: | 2006-03-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants of gating at the potassium-channel selectivity filter.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ATK
 
 | | Structure of a mutant KcsA K+ channel | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, NONAN-1-OL, ... | | Authors: | Cordero-Morales, J.F, Cuello, L.G, Zhao, Y, Jogini, V, Chakrapani, S, Roux, B, Perozo, E. | | Deposit date: | 2005-08-25 | | Release date: | 2006-03-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants of gating at the potassium-channel selectivity filter.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6V1X
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Tetramer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
6V1Y
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Octamer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
5VKH
 
 | | Closed conformation of KcsA Y82A-F103A mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Cuello, L.G, Perozo, E, Cortes, D.M. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The gating cycle of a K+ channel at atomic resolution.
Elife, 6, 2017
|
|
5VK6
 
 | | Open conformation of KcsA non-inactivating E71A mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Cuello, L.G, Perozo, E. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The gating cycle of a K+ channel at atomic resolution.
Elife, 6, 2017
|
|
5VKE
 
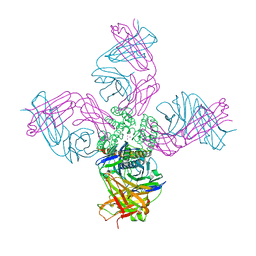 | | Open conformation of KcsA deep-inactivated | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Cuello, L.G, Perozo, E, Cortes, D.M. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The gating cycle of a K+ channel at atomic resolution.
Elife, 6, 2017
|
|
6PWN
 
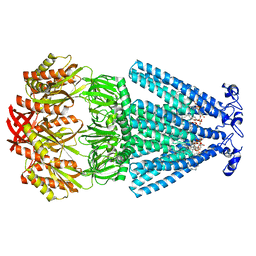 | | MscS Nanodisc with N-terminal His-Tag | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, HEXADECANE, Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
6PWO
 
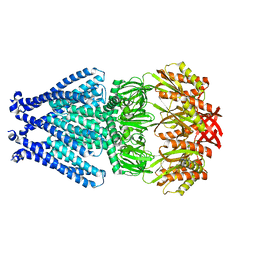 | | MscS DDM | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
6PWP
 
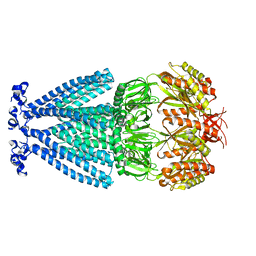 | | MscS Nanodisc | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
3STZ
 
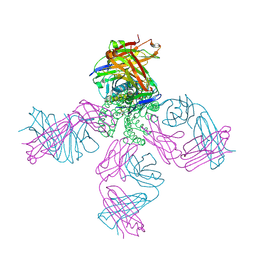 | | KcsA potassium channel mutant Y82C with nitroxide spin label | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
3STL
 
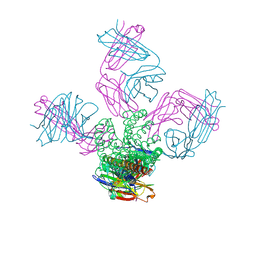 | | KcsA potassium channel mutant Y82C with Cadmium bound | | Descriptor: | CADMIUM ION, POTASSIUM ION, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
2P7T
 
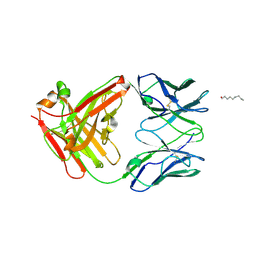 | | Crystal Structure of KcsA mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB-A, FAB-B, ... | | Authors: | Cordero-Morales, J.F, Vishwanath, J, Lewis, A, Valeria, V.R, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-10-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of KcsA mutant
To be Published
|
|
2QI9
 
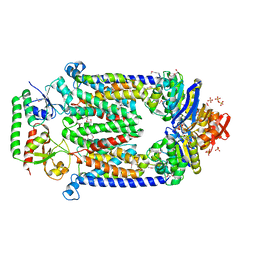 | | ABC-transporter BtuCD in complex with its periplasmic binding protein BtuF | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PHOSPHATE ION, ... | | Authors: | Hvorup, R.N, Goetz, B.A, Niederer, M, Hollenstein, K, Perozo, E, Locher, K.P. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF.
Science, 317, 2007
|
|
7S9C
 
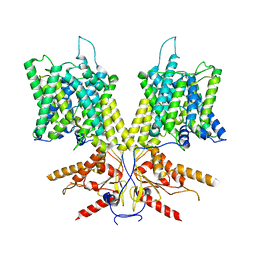 | | Cryo-EM Structure of dolphin Prestin: Sensor Down II (Expanded II) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9B
 
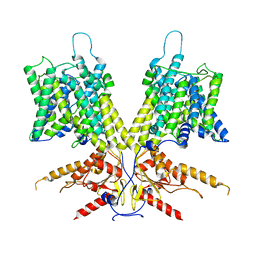 | | Cryo-EM Structure of dolphin Prestin: Sensor Down I (Expanded) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9D
 
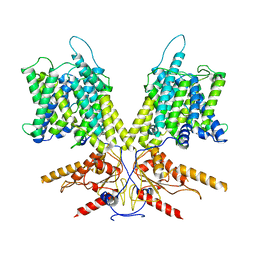 | | Cryo-EM Structure of dolphin Prestin: Intermediate state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9E
 
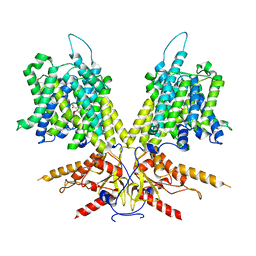 | | Cryo-EM Structure of dolphin Prestin: Inhibited II (Sulfate +Salicylate) state | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9A
 
 | | Cryo-EM Structure of dolphin Prestin: Inhibited I (Chloride + Salicylate) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S8X
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Up (compact) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
