1M3J
 
 | |
1M3I
 
 | | Perfringolysin O, new crystal form | | Descriptor: | perfringolysin O | | Authors: | Rossjohn, J, Parker, M, Polekhina, G, Feil, S, Tweten, R. | | Deposit date: | 2002-06-28 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Snapshots in the Molecular Mechanism of PFO Revealed
To be Published
|
|
1OQX
 
 | | G-2 glycovariant of human IgG Fc bound to minimized version of Protein A called Z34C | | Descriptor: | Protein A Z34C, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Raju, T.S, Mulkerrin, M.G, Parker, M, De Vos, A.M, Gazzano-Santoro, H, Totpal, K, Ultsch, M.H. | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of Fc Glycans on The Effector Functions Vary
with the Antibody mechanism of Action
To be Published
|
|
1OQO
 
 | | Complex between G0 version of an Fc bound to a minimized version of Protein A called Mini-Z | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Minimized version of Protein A (Z34C), ... | | Authors: | Raju, T.S, Mulkerrin, M.G, Parker, M, De Vos, A.M, Gazzano-Santoro, H, Totpal, K, Ultsch, M.H. | | Deposit date: | 2003-03-10 | | Release date: | 2003-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of Fc Glycans on The Effector
Functions Vary with the Antibody
mechanism of Action
To be Published
|
|
10GS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
9GSS
 
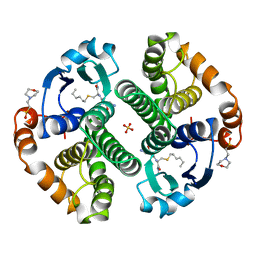 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH S-HEXYL GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, S-HEXYLGLUTATHIONE, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
8GSS
 
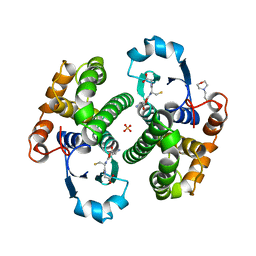 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
4GSS
 
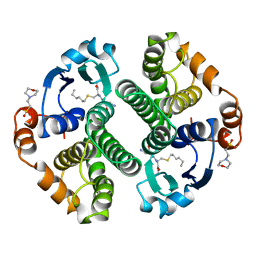 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y108F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A, Rossjohn, J, Parker, M. | | Deposit date: | 1997-01-20 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme.
Biochemistry, 36, 1997
|
|
5GSS
 
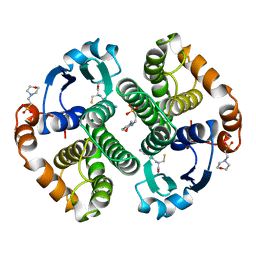 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
7GSS
 
 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
6GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
3TZR
 
 | | Structure of a Riboswitch-like RNA-ligand complex from the Hepatitis C Virus Internal Ribosome Entry Site | | Descriptor: | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine, 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3', 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3', ... | | Authors: | Dibrov, S.M, Ding, K, Brunn, N, Parker, M.A, Bergdahl, B.M, Wyles, D.L, Hermann, T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
To be Published
|
|
3ZXZ
 
 | | X-ray Structure of PF-04217903 bound to the kinase domain of c-Met | | Descriptor: | 2-{4-[1-(QUINOLIN-6-YLMETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRAZIN-6-YL]-1H-PYRAZOL-1-YL}ETHANOL, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3ZZE
 
 | | Crystal structure of C-MET kinase domain in complex with N'-((3Z)-4- chloro-7-methyl-2-oxo-1,2-dihydro-3H-indol-3-ylidene)-2-(4- hydroxyphenyl)propanohydrazide | | Descriptor: | (2S)-N'-[(3R)-4-chloro-7-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-2-(4-hydroxyphenyl)propanehydrazide, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Deng, Y, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4AP7
 
 | | Crystal structure of C-MET kinase domain in complex with 4-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl)phenol | | Descriptor: | 4-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]phenol, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4AOI
 
 | | Crystal structure of C-MET kinase domain in complex with 4-(3-((1H- pyrrolo(2,3-b)pyridin-3-yl)methyl)-(1,2,4)triazolo(4,3-b)(1,2,4) triazin-6-yl)benzonitrile | | Descriptor: | 4-[3-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-6-yl]benzenecarbonitrile, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3ZC5
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor (S)-6-(1-(6- (1-methyl-1H-pyrazol-4-yl)-(1,2,4)triazolo(4,3-b)pyridazin-3-yl)ethyl) quinoline. | | Descriptor: | 6-{(1S)-1-[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]ethyl}quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
3ZCL
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor (S)-3-(1-(1H-pyrrolo(2,3-b)pyridin-3-yl)ethyl)-N-isopropyl-(1,2,4)triazolo(4,3- b)pyridazin-6-amine | | Descriptor: | (S)-3-(1-(1H-pyrrolo(2,3-b)pyridin-3-yl)ethyl)-N-isopropyl-(1,2,4)triazolo(4,3-b)pyridazin-6-amine, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
3ZBX
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor 6-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl) quinoline. | | Descriptor: | 6-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J, Ryan, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
2LU3
 
 | |
2LU4
 
 | |
5C37
 
 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | Descriptor: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | Authors: | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of a Series of Bioavailable Fatty Acid Synthase (FASN) KR Domain Inhibitors for Cancer Therapy
to be published
|
|
