5CJ9
 
 | | Bacillus halodurans Arginine repressor, ArgR | | Descriptor: | Arginine repressor, S-1,2-PROPANEDIOL | | Authors: | Park, Y.W, Kang, J, Yeo, H.Y, Lee, J.Y. | | Deposit date: | 2015-07-14 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Structural Analysis and Insights into the Oligomeric State of an Arginine-Dependent Transcriptional Regulator from Bacillus halodurans.
Plos One, 11, 2016
|
|
5ZZ6
 
 | | Redox-sensing transcriptional repressor Rex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor Rex 1 | | Authors: | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-05-30 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
5ZZ7
 
 | | Redox-sensing transcriptional repressor Rex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Redox-sensing transcriptional repressor Rex 1 | | Authors: | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-05-30 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
5ZZ5
 
 | | Redox-sensing transcriptional repressor Rex | | Descriptor: | GLYCEROL, Redox-sensing transcriptional repressor Rex | | Authors: | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-05-30 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
7WB3
 
 | | Crystal structure of T. maritima Rex in ternary complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*GP*AP*GP*AP*AP*AP*TP*TP*TP*AP*TP*CP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*TP*GP*AP*TP*AP*AP*AP*TP*TP*TP*CP*TP*CP*AP*AP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lee, J.Y, Jeong, K.H, Lee, H.J, Park, Y.W. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA.
Int J Mol Sci, 23, 2022
|
|
5H38
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | ORF70, PHOSPHATE ION | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5GPA
 
 | |
5H3A
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70, TOMUDEX | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5H39
 
 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70 | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5GP9
 
 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | GLYCEROL, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Lee, J.Y, Yeo, H.K, Park, Y.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
7CV2
 
 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | Descriptor: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
4Q16
 
 | |
5GPC
 
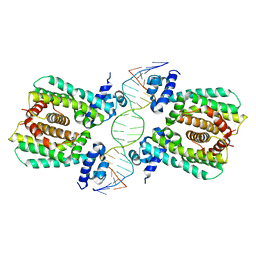 | | Structural analysis of fatty acid degradation regulator FadR from Bacillus halodurans | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*AP*TP*GP*AP*GP*TP*AP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), DNA (5'-D(P*GP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*AP*CP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Transcriptional regulator (TetR/AcrR family) | | Authors: | Lee, J.Y, Yeo, H.K, Park, T.W. | | Deposit date: | 2016-08-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR.
Nucleic Acids Res., 45, 2017
|
|
3S22
 
 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with an inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, [(2S,3R)-2-formyl-1-{[4-(methylamino)butyl]carbamoyl}pyrrolidin-3-yl]sulfamic acid | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3S1Y
 
 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4NK3
 
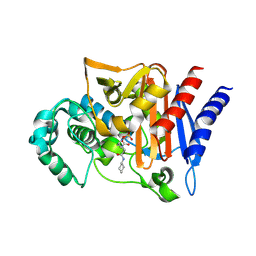 | | Amp-c beta-lactamase (pseudomonas aeruginosa) in complex with mk-7655 | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin().
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O5V
 
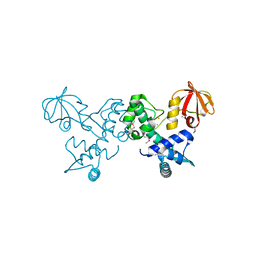 | | Crystal structure of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O6J
 
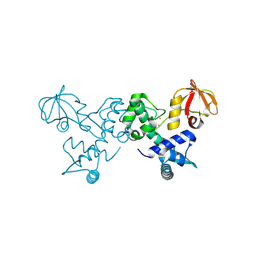 | | Crystal sturucture of T. acidophilum IdeR | | Descriptor: | FE (II) ION, Iron-dependent transcription repressor related protein | | Authors: | Lee, J.Y, Yeo, H.K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5ZLP
 
 | | Crystal structure of glutamine synthetase from helicobacter pylori | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Joo, H.K, Lee, J.Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Analysis of Glutamine Synthetase from Helicobacter pylori.
Sci Rep, 8, 2018
|
|
5ZLI
 
 | |
1MOL
 
 | |
7CHT
 
 | | Crystal structure of TTK kinase domain in complex with compound 30 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(oxan-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Ko, E.H, Choi, H.G, Son, J.B, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CHM
 
 | | Crystal structure of TTK kinase domain in complex with compound 8 | | Descriptor: | 4-(cyclohexylamino)-2-[(2-methoxy-4-morpholin-4-ylcarbonyl-phenyl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7CHN
 
 | | Crystal structure of TTK kinase domain in complex with compound 9 | | Descriptor: | 4-(cyclohexylamino)-2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
