5CSR
 
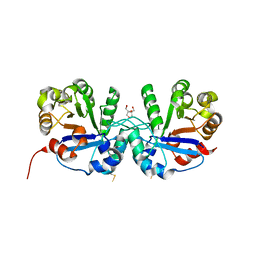 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilium | | Descriptor: | CHLORIDE ION, GLYCEROL, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Park, H.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
3LJ8
 
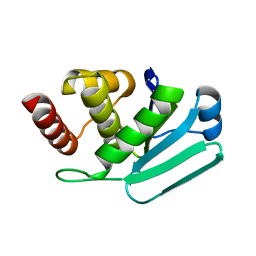 | | Crystal Structure of MKP-4 | | Descriptor: | Tyrosine-protein phosphatase | | Authors: | Jeong, D.G, Yoon, T.S, Jung, S.-K, Park, H.S, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring binding sites other than the catalytic core in the crystal structure of the catalytic domain of MKP-4
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2OS1
 
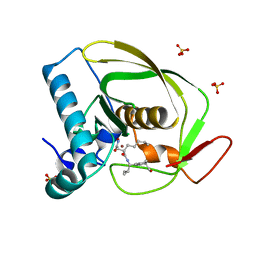 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS0
 
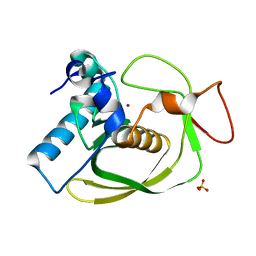 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
5CSS
 
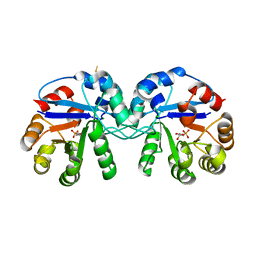 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
1PLY
 
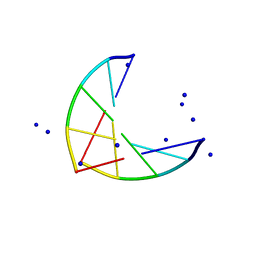 | | SODIUM IONS AND WATER MOLECULES IN THE STRUCTURE OF POLY D(A)(DOT)POLY D(T) | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*T)-3'), SODIUM ION | | Authors: | Chandrasekaran, R, Radha, A, Park, H.-S. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (3.2 Å) | | Cite: | Sodium ions and water molecules in the structure of poly(dA).poly(dT).
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2OS3
 
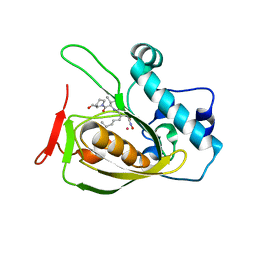 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Parh, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
7XGE
 
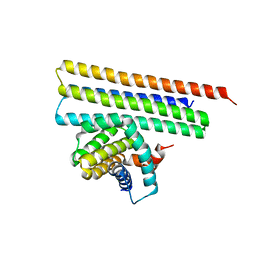 | |
7XGF
 
 | |
7XGG
 
 | |
5ZOO
 
 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP1 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP1 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
5ZOP
 
 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP2 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP2 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
3GWJ
 
 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4N6R
 
 | | Crystal structure of VosA-VelB-complex | | Descriptor: | SULFATE ION, VelB, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
2ODR
 
 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
