8I2N
 
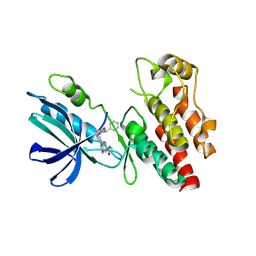 | | The RIPK1 kinase domain in complex with QY7-2B compound | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-methyl-1-[4-[[[1-methyl-5-(phenylmethyl)pyrazol-3-yl]carbonylamino]methyl]phenyl]benzimidazole-5-carboxamide | | Authors: | Gong, X.Y, Li, Y, Meng, H.Y, Pan, L.F. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The RIPK1 kinase domain in complex with QY7-2B compound
To Be Published
|
|
7C7J
 
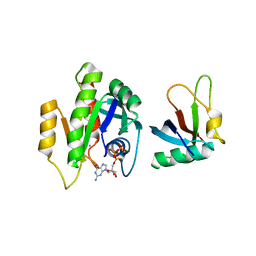 | |
7C7I
 
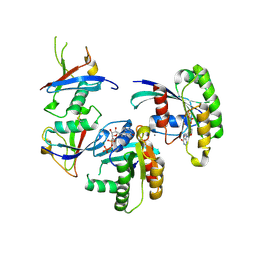 | |
7CWW
 
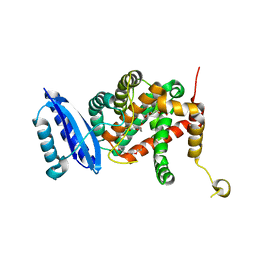 | | Crystal structure of TsrL | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, TsrE | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-08-31 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of TsrL
To Be Published
|
|
5Z7A
 
 | | Crystal structure of NDP52 SKICH region | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q. | | Deposit date: | 2018-01-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z7G
 
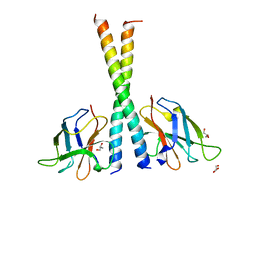 | | Crystal structure of TAX1BP1 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Tax1-binding protein 1 | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q, Wang, Y.L, Hu, S.C. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8JC5
 
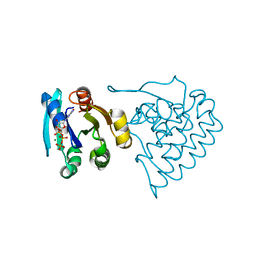 | |
8JCA
 
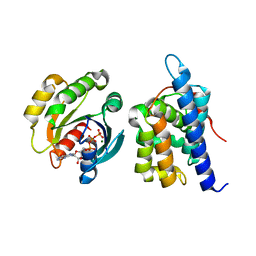 | |
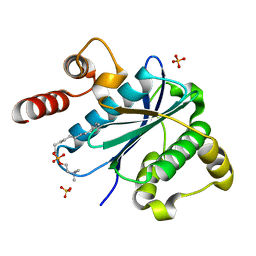
3KYQ
 
 | |
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
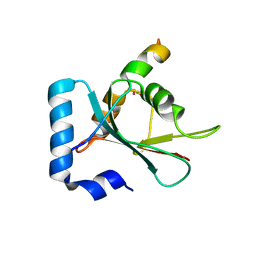 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7D0E
 
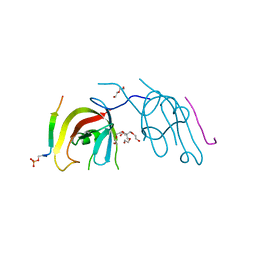 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZM
 
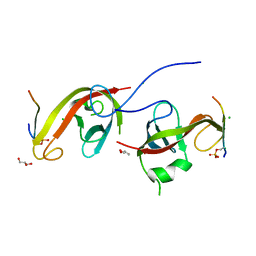 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZG
 
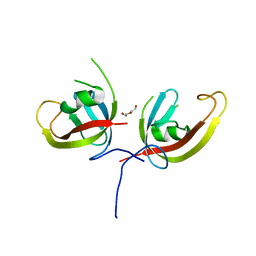 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
5Z7L
 
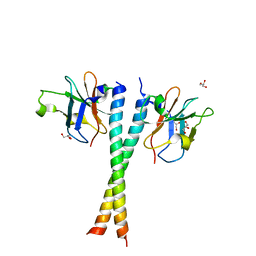 | | Crystal structure of NDP52 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL | | Authors: | Fu, T, Pan, L.F. | | Deposit date: | 2018-01-29 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3PVL
 
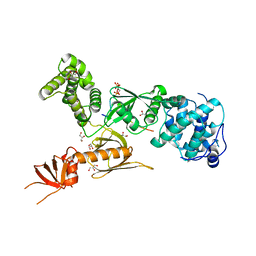 | | Structure of myosin VIIa MyTH4-FERM-SH3 in complex with the CEN1 of Sans | | Descriptor: | GLYCEROL, Myosin VIIa isoform 1, PHOSPHATE ION, ... | | Authors: | Wu, L, Pan, L.F, Wei, Z.Y, Zhang, M.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo.
Science, 331, 2011
|
|
7X80
 
 | |
7X7Z
 
 | |
7X86
 
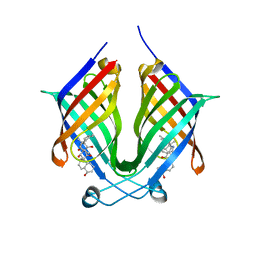 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X81
 
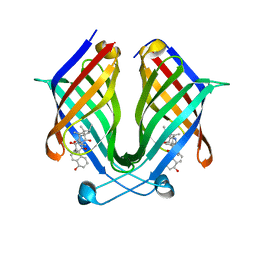 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7XFR
 
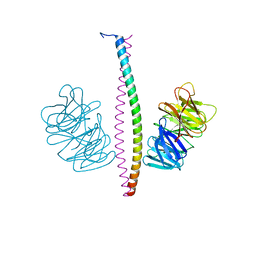 | |
7YJ0
 
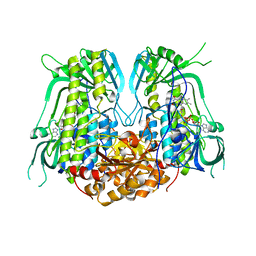 | |
6KKG
 
 | | Crystal structure of MAGI2-Dendrin complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, Peptide from Dendrin | | Authors: | Zhu, J.W, Zhang, H.J, Lin, L, Zhang, R.G. | | Deposit date: | 2019-07-25 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Phase separation of MAGI2-mediated complex underlies formation of slit diaphragm complex in glomerular filtration barrier
To Be Published
|
|
7F69
 
 | | Crystal structure of WIPI2b in complex with ATG16L1 | | Descriptor: | Isoform 2 of Autophagy-related protein 16-1, Isoform 2 of WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Gong, X.Y, Pan, L.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy.
Sci Adv, 9, 2023
|
|
