3BL2
 
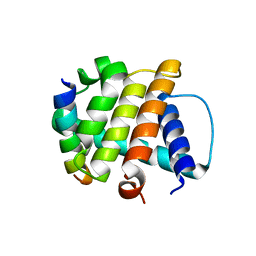 | | Crystal Structure of M11, the BCL-2 Homolog of Murine Gamma-herpesvirus 68, Complexed with Mouse Beclin1 (residues 106-124) | | Descriptor: | Beclin-1, V-bcl-2 | | Authors: | Oh, B.-H, Woo, J.-S, Ku, B. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine gamma-Herpesvirus 68
Plos Pathog., 4, 2008
|
|
1DU3
 
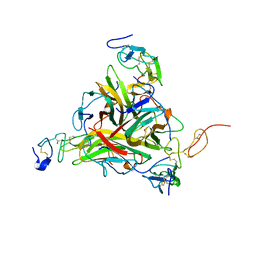 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
3ZTH
 
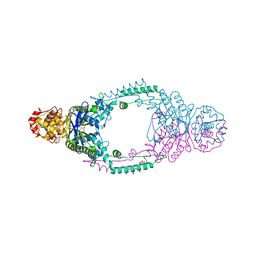 | | Crystal structure of Stu0660 of Streptococcus thermophilus | | Descriptor: | MAGNESIUM ION, STU0660 | | Authors: | Lee, K.-H, Robinson, H, Oh, B.-H. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, Structural, and Biochemical Characterization of a Group of Large Csn2 Proteins Involved in Crispr-Mediated Bacterial Immunity.
Proteins, 80, 2012
|
|
4AKF
 
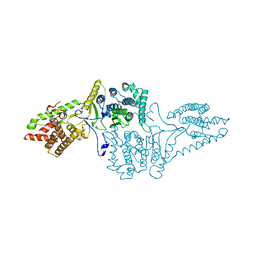 | |
2J3R
 
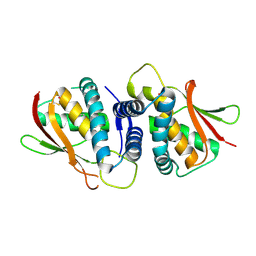 | | The crystal structure of the bet3-trs31 heterodimer. | | Descriptor: | NITRATE ION, PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, ... | | Authors: | Kim, Y.-G, Oh, B.-H. | | Deposit date: | 2006-08-23 | | Release date: | 2006-11-27 | | Last modified: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Cell(Cambridge,Mass.), 127, 2006
|
|
2J3W
 
 | | The crystal structure of the bet3-trs31-sedlin complex. | | Descriptor: | PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX PROTEIN 2, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, ... | | Authors: | Kim, Y.-G, Oh, B.-H. | | Deposit date: | 2006-08-23 | | Release date: | 2006-11-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Cell(Cambridge,Mass.), 127, 2006
|
|
2WCV
 
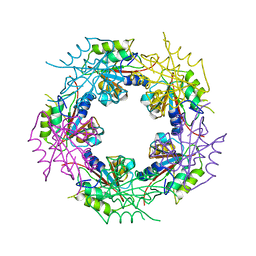 | | Crystal structure of bacterial FucU | | Descriptor: | L-FUCOSE MUTAROTASE, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
2WCU
 
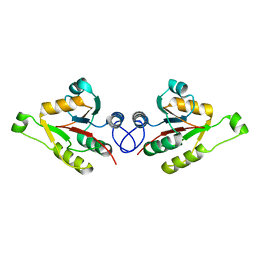 | | Crystal structure of mammalian FucU | | Descriptor: | PROTEIN FUCU HOMOLOG, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
6JUV
 
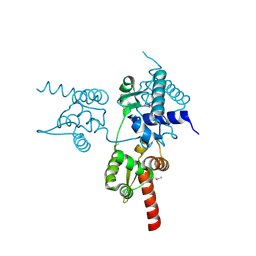 | |
2C0J
 
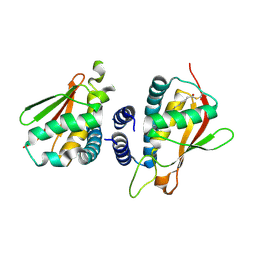 | | Crystal structure of the bet3-trs33 heterodimer | | Descriptor: | PALMITIC ACID, R32611_2, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3 | | Authors: | Kim, M.-S, Yi, M.-J, Lee, K.-H, Wagner, J, Munger, C, Kim, Y.-G, Whiteway, M, Cygler, M, Oh, B.-H, Sacher, M. | | Deposit date: | 2005-09-03 | | Release date: | 2006-02-07 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies Reveal a Specific Interaction between Trapp Subunits Trs33P and Bet3P
Traffic, 6, 2005
|
|
2CB3
 
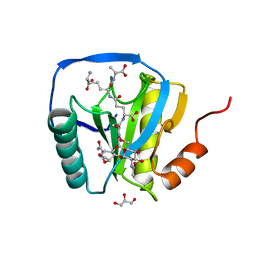 | | Crystal structure of peptidoglycan recognition protein-LE in complex with tracheal cytotoxin (monomeric diaminopimelic acid-type peptidoglycan) | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, GLYCEROL, PEPTIDOGLYCAN-RECOGNITION PROTEIN-LE | | Authors: | Lim, J.-H, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2005-12-29 | | Release date: | 2006-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Preferential Recognition of Diaminopimelic Acid-Type Peptidoglycan by a Subset of Peptidoglycan Recognition Proteins
J.Biol.Chem., 281, 2006
|
|
1E97
 
 | |
6IVH
 
 | |
5XEI
 
 | |
5XG3
 
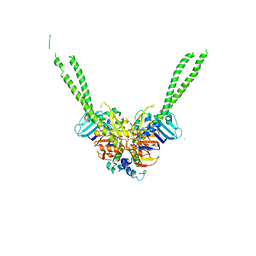 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XG2
 
 | |
5XNS
 
 | |
1V0D
 
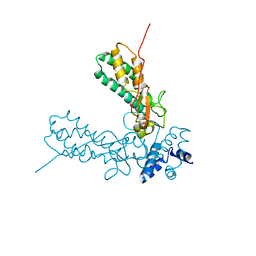 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
1W6Y
 
 | | crystal structure of a mutant W92A in ketosteroid isomerase (KSI) from Pseudomonas putida biotype B | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Yun, Y.S, Nam, G.H, Kim, Y.-G, Oh, B.-H, Choi, K.Y. | | Deposit date: | 2004-08-25 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Exterior Hydrophobic Cluster Contributes to Conformational Stability and Steroid Binding in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
FEBS J., 272, 2005
|
|
1WC8
 
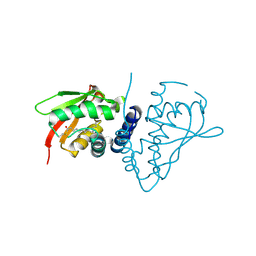 | | The crystal structure of mouse bet3p | | Descriptor: | MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | Authors: | Kim, Y.-G, Sacher, M, Oh, B.-H. | | Deposit date: | 2004-11-10 | | Release date: | 2004-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WC9
 
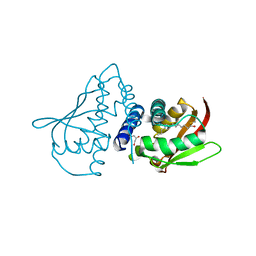 | | The crystal structure of truncated mouse bet3p | | Descriptor: | GLYCEROL, MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | Authors: | Kim, Y.-G, Lee, H.-S, Sacher, M, Oh, B.-H. | | Deposit date: | 2004-11-10 | | Release date: | 2004-12-13 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
1QJG
 
 | |
1GS3
 
 | |
1E9Z
 
 | |
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
