4LH8
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
3H96
 
 | | Msmeg_3358 F420 Reductase | | Descriptor: | 1,2-ETHANEDIOL, F420-H2 Dependent Reductase A | | Authors: | Jackson, C.J, French, N, Newman, J, Taylor, M.C, Russell, R.J, Oakeshott, J.G. | | Deposit date: | 2009-04-30 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation.
Mol.Microbiol., 78, 2010
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4FNG
 
 | | The alpha-esterase-7 carboxylesterase, E3, from the blowfly Lucilia cuprina | | Descriptor: | E3 alpha-esterase-7 caboxylesterase | | Authors: | Jackson, C.J, Liu, J.-W, Carr, P.D, Younis, F, Pandey, G, Coppin, C, Meirelles, T, Ollis, D.L, Tawfik, D.S, Weik, M, Oakeshott, J.G. | | Deposit date: | 2012-06-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of an insect alpha-carboxylesterase ( alpha Esterase7) associated with insecticide resistance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FNM
 
 | | The alpha-esterase-7 carboxylesterase, E3, from the blowfly Lucilia cuprina | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 alpha-esterase-7 carboxylesterase | | Authors: | Jackson, C.J, Liu, J.-W, Carr, P.D, Younis, F, Pandey, G, Coppin, C, Meirelles, T, Ollis, D.L, Tawfik, D.S, Weik, M, Oakeshott, J.G. | | Deposit date: | 2012-06-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and function of an insect alpha-carboxylesterase ( alpha Esterase7) associated with insecticide resistance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2O7V
 
 | | Carboxylesterase AeCXE1 from Actinidia eriantha covalently inhibited by paraoxon | | Descriptor: | CXE carboxylesterase, DIETHYL PHOSPHONATE | | Authors: | Ileperuma, N.R, Marshall, S.D, Squire, C.J, Baker, H.M, Oakeshott, J.G, Russell, R.J, Plummer, K.M, Newcomb, R.D, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-Resolution Crystal Structure of Plant Carboxylesterase AeCXE1, from Actinidia eriantha, and Its Complex with a High-Affinity Inhibitor Paraoxon.
Biochemistry, 46, 2007
|
|
2O7R
 
 | | Plant carboxylesterase AeCXE1 from Actinidia eriantha with acyl adduct | | Descriptor: | CXE carboxylesterase, PROPYL ACETATE | | Authors: | Ileperuma, N.R, Marshall, S.D, Squire, C.J, Baker, H.M, Oakeshott, J.G, Russell, R.J, Plummer, K.M, Newcomb, R.D, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Crystal Structure of Plant Carboxylesterase AeCXE1, from Actinidia eriantha, and Its Complex with a High-Affinity Inhibitor Paraoxon.
Biochemistry, 46, 2007
|
|
4WHB
 
 | | Crystal structure of phenylurea hydrolase B | | Descriptor: | Phenylurea hydrolase B, ZINC ION | | Authors: | Sugrue, E, Carr, P.D, Khurana, J.L, Jackson, C.J. | | Deposit date: | 2014-09-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
4WGX
 
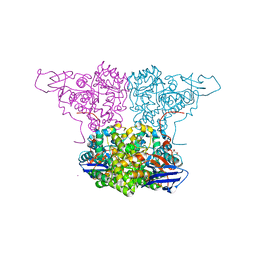 | | Crystal Structure of Molinate Hydrolase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT (II) ION, Molinate hydrolase | | Authors: | Sugrue, E, Carr, P.D, Fraser, N.J, Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
3F7E
 
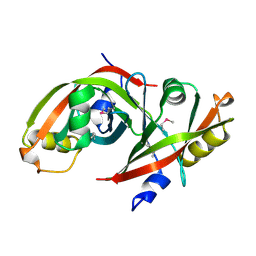 | | MSMEG_3380 F420 Reductase | | Descriptor: | Pyridoxamine 5'-phosphate oxidase-related, FMN-binding | | Authors: | Jackson, C.J. | | Deposit date: | 2008-11-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Identification and characterisation of two novel classes of F420-dependent reductases from Mycobacterium that degrade aflatoxin
To be Published
|
|
5THM
 
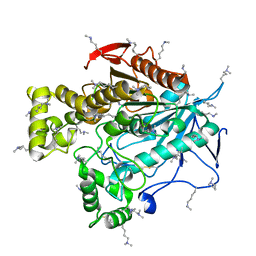 | | Esterase-6 from Drosophila melanogaster | | Descriptor: | Esterase-6, N,N-dimethylboranamine | | Authors: | Fraser, N.J, Jackson, C.J. | | Deposit date: | 2016-09-29 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the behavioral effects of the odorant degrading enzyme Esterase 6 in Drosophila.
Sci Rep, 7, 2017
|
|
5W1U
 
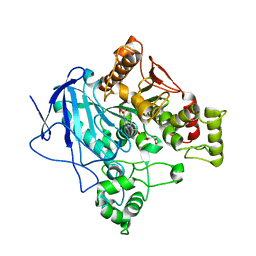 | | Culex quinquefasciatus carboxylesterase B2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carboxylic ester hydrolase, MALONATE ION | | Authors: | Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Insecticide Sequestering Carboxylesterase from the Disease Vector Culex quinquefasciatus: What Makes an Enzyme a Good Insecticide Sponge?
Biochemistry, 56, 2017
|
|
5IKX
 
 | |
6UW1
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UVX
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Apo state | | Descriptor: | CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW5
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, GDP and Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW7
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, Dehydro-F420-0 bound form | | Descriptor: | 2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxyprop-2-enoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Izore, T, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW3
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
