8SEM
 
 | |
8SED
 
 | |
1D1F
 
 | |
1D1E
 
 | |
1D0W
 
 | |
5TBD
 
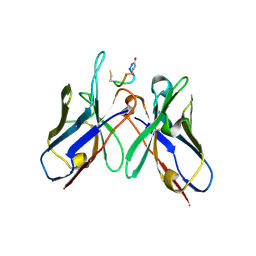 | | Crystal Structure of anti-MSP2 Fv fragment (mAb4D11) in complex with 3D7-MSP2 215-222 | | Descriptor: | Fv fragment (mAb4D1) heavy chain, Merozoite surface protein 2 | | Authors: | Seow, J, Morales, R.A.V, MacRaild, C.A, Bankala, K, Drinkwater, N, Dingjan, T, Jaipuria, G, Wilde, K, Anders, R.F, Atreya, H.S, Christ, D, McGowan, S, Norton, R.S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
J. Mol. Biol., 429, 2017
|
|
1AHL
 
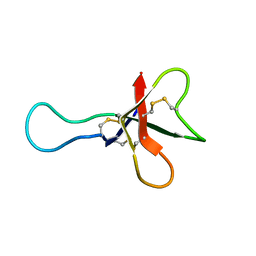 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
1VIB
 
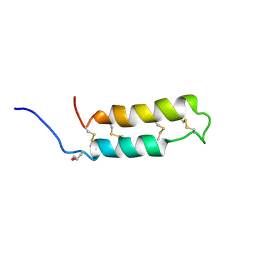 | | NMR SOLUTION STRUCTURE OF THE NEUROTOXIN B-IV, 20 STRUCTURES | | Descriptor: | NEUROTOXIN B-IV | | Authors: | Barnham, K.J, Dyke, T.R, Kem, W.R, Norton, R.S. | | Deposit date: | 1996-11-25 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding.
J.Mol.Biol., 268, 1997
|
|
8F04
 
 | |
1APF
 
 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
3EK9
 
 | | SPRY Domain-containing SOCS Box Protein 2: Crystal Structure and Residues Critical for Protein Binding | | Descriptor: | GLYCEROL, SPRY domain-containing SOCS box protein 2 | | Authors: | Kuang, Z, Yao, S, Xu, Y, Garrett, T.J.P, Norton, R.S. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding.
J.Mol.Biol., 386, 2009
|
|
6MJV
 
 | | A consensus human beta defensin | | Descriptor: | Human beta-defensin | | Authors: | Amero, C, Villegas-Moreno, J, Rodriguez, A, Norton, R.S, Corzo, G. | | Deposit date: | 2018-09-22 | | Release date: | 2019-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial activity and structure of a consensus human beta-defensin and its comparison to a novel putative hBD10.
Proteins, 88, 2020
|
|
1ANS
 
 | |
6OQP
 
 | | U-AITx-Ate1 | | Descriptor: | SER-LYS-TRP-ILE-CYS-ALA-ASN-ARG-SER-VAL-CYS-PRO-ILE | | Authors: | Elnahriry, K.A, Wai, D.C.C, Norton, R.S. | | Deposit date: | 2019-04-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterisation of a novel peptide from the Australian sea anemone Actinia tenebrosa.
Toxicon, 168, 2019
|
|
2SH1
 
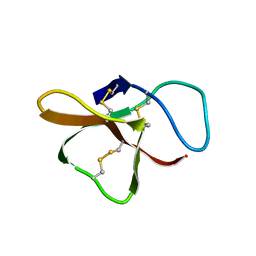 | |
1IGL
 
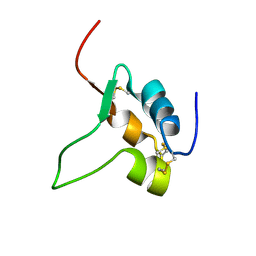 | | SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR II RELATIONSHIP TO RECEPTOR AND BINDING PROTEIN INTERACTIONS | | Descriptor: | INSULIN-LIKE GROWTH FACTOR II | | Authors: | Torres, A.M, Forbes, B.E, Aplin, S.E, Wallace, J.C, Francis, G.L, Norton, R.S. | | Deposit date: | 1994-12-29 | | Release date: | 1995-02-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions.
J.Mol.Biol., 248, 1995
|
|
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | Descriptor: | spasmodic protein tx9a-like protein | | Authors: | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2002-07-04 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
2AFJ
 
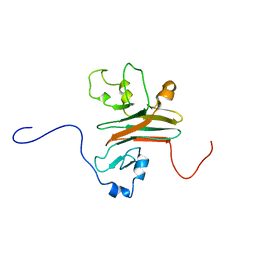 | | SPRY domain-containing SOCS box protein 2 (SSB-2) | | Descriptor: | gene rich cluster, C9 gene | | Authors: | Masters, S.L, Yao, S, Willson, T.A, Zhang, J.G, Palmer, K.R, Smith, B.J, Babon, J.J, Nicola, N.A, Norton, R.S, Nicholson, S.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-01-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues
Nat.Struct.Mol.Biol., 13, 2006
|
|
2BBU
 
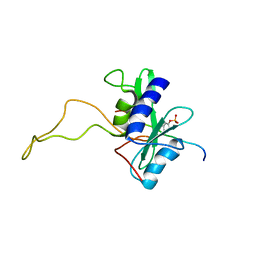 | |
2CCO
 
 | |
1YXE
 
 | | Structure and inter-domain interactions of domain II from the blood stage malarial protein, apical membrane antigen 1 | | Descriptor: | apical membrane antigen 1 | | Authors: | Feng, Z.P, Keizer, D.W, Stevenson, R.A, Yao, S, Babon, J.J, Murphy, V.J, Anders, R.F, Norton, R.S. | | Deposit date: | 2005-02-21 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Inter-domain Interactions of Domain II from the Blood-stage Malarial Protein, Apical Membrane Antigen 1.
J.Mol.Biol., 350, 2005
|
|
1ZEC
 
 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
5JYQ
 
 | | Structure of Conus Geographus insulin G1 | | Descriptor: | Insulin 1, Insulin 1b, SULFATE ION | | Authors: | Lawrence, M.C, Menting, J, Norton, R.S, Safavi-Hemami, H. | | Deposit date: | 2016-05-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4R1C
 
 | |
