1TER
 
 | |
5KUY
 
 | |
5KUX
 
 | |
4J8T
 
 | | Engineered Digoxigenin binder DIG10.2 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.2 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4J9A
 
 | | Engineered Digoxigenin binder DIG10.3 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.3 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
7KL9
 
 | |
5T83
 
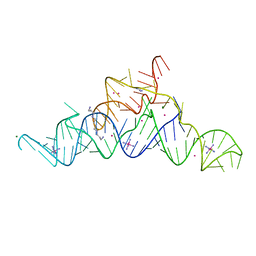 | | Structure of a guanidine-I riboswitch from S. acidophilus | | Descriptor: | GUANIDINE, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Reiss, C.W, Xiong, Y, Strobel, S.A. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Ligand Binding to the Guanidine-I Riboswitch.
Structure, 25, 2017
|
|
