3G66
 
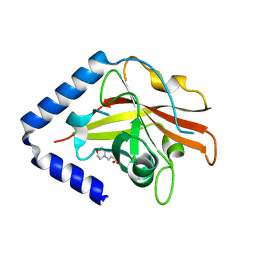 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
3G69
 
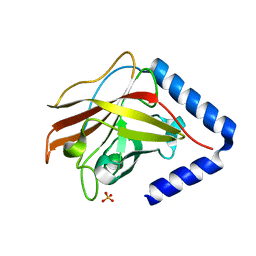 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
6YAW
 
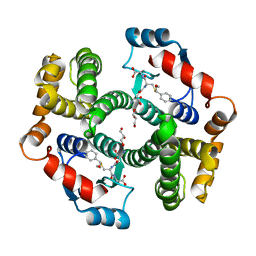 | | Crystal structure of human GSTA1-1 bound to the glutathione adduct of cinnamaldehyde | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-[(1~{R})-3-oxidanylidene-1-phenyl-propyl]sulfanyl-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, GLYCEROL, Glutathione S-transferase A1 | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2020-03-13 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions Between Odorants and Glutathione Transferases in the Human Olfactory Cleft.
Chem.Senses, 45, 2020
|
|
8Q8B
 
 | |
8Q89
 
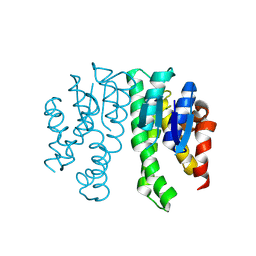 | |
8Q8A
 
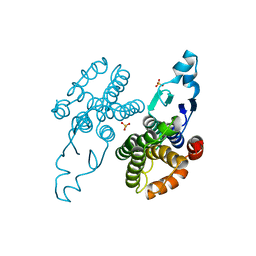 | |
8BB8
 
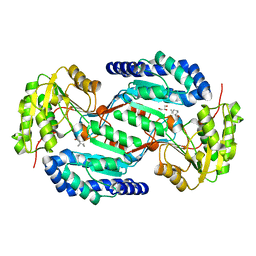 | |
6GFA
 
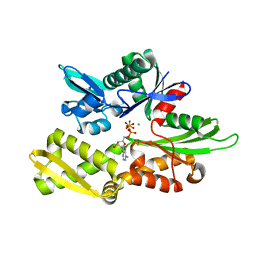 | | Structure of Nucleotide binding domain of HSP110, ATP and Mg2+ complexed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 105 kDa, MAGNESIUM ION | | Authors: | Gonzalez, D, Gotthard, G, Gozzi, G.J, Seigneuric, R, Neiers, F, Briand, L, Garrido, C. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy.
Cell Death Differ., 27, 2020
|
|
5F0G
 
 | |
5OLL
 
 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
7BIC
 
 | |
7BIA
 
 | | Crystal structure of human GSTP1 bound to iberin | | Descriptor: | 1-isothiocyanato-3-methylsulfinyl-propane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, ... | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Role of human salivary enzymes in bitter taste perception.
Food Chem, 386, 2022
|
|
7BIB
 
 | |
6QQ4
 
 | | Odorant-binding protein dmelOBP28a from Drosophila melanogaster | | Descriptor: | General odorant-binding protein 28a, MALONIC ACID, PENTAETHYLENE GLYCOL | | Authors: | Gonzalez, D, Neiers, F, Gotthard, G, Briand, L. | | Deposit date: | 2019-02-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The Drosophila odorant-binding protein 28a is involved in the detection of the floral odour ss-ionone.
Cell.Mol.Life Sci., 77, 2020
|
|
6T2T
 
 | | Crystal structure of Drosophila melanogaster glutathione S-transferase epsilon 14 in complex with glutathione and 2-methyl-2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Skerlova, J, Lindstrom, H, Sjodin, B, Gonis, E, Neiers, F, Mannervik, B, Stenmark, P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and steroid isomerase activity of Drosophila glutathione transferase E14 essential for ecdysteroid biosynthesis.
Febs Lett., 594, 2020
|
|
2JZS
 
 | | Solution structure of the reduced form of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2JZR
 
 | | Solution structure of the oxidized form (Cys67-Cys70) of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2FY6
 
 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
7QUG
 
 | |
3HCG
 
 | |
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCJ
 
 | |
3HCI
 
 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
8AWZ
 
 | |
8AX2
 
 | |
