1AMH
 
 | | UNCOMPLEXED RAT TRYPSIN MUTANT WITH ASP 189 REPLACED WITH SER (D189S) | | Descriptor: | ANIONIC TRYPSIN, CALCIUM ION | | Authors: | Szabo, E, Bocskei, Z.S, Naray-Szabo, G, Graf, L. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of Asp189Ser trypsin provides evidence for an inherent structural plasticity of the protease.
Eur.J.Biochem., 263, 1999
|
|
1QIV
 
 | | CALMODULIN COMPLEXED WITH N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE (DPD), 1:2 COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Naray-Szabo, G, Ovadi, J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
1A29
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:2 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Bocskei, Zs, Harmat, V, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1998-01-19 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies.
Biochemistry, 37, 1998
|
|
1ZJK
 
 | | Crystal structure of the zymogen catalytic region of human MASP-2 | | Descriptor: | Mannan-binding lectin serine protease 2 | | Authors: | Gal, P, Harmat, V, Kocsis, A, Bian, T, Barna, L, Ambrus, G, Vegh, B, Balczer, J, Sim, R.B, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2005-04-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A True Autoactivating Enzyme: Structural insight into mannose-binding lectin-associated serine protease-2 activations
J.Biol.Chem., 280, 2005
|
|
3EQ7
 
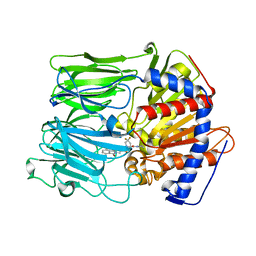 | | Prolyl oligopeptidase complexed with R-Pro-(decarboxy-Pro)-Type inhibitors | | Descriptor: | 2-{3-[(2S)-4,4-difluoro-2-(pyrrolidin-1-ylcarbonyl)pyrrolidin-1-yl]-3-oxopropyl}-isoindole-1,3(2H)-dione, Prolyl endopeptidase | | Authors: | Kanai, K, Aranyi, P, Bocskei, Z, Ferenczy, G, Harmat, V, Simon, K, Naray-Szabo, G, Hermecz, I. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Prolyl oligopeptidase inhibition by N-acyl-pro-pyrrolidine-type molecules
J.Med.Chem., 51, 2008
|
|
3EQ8
 
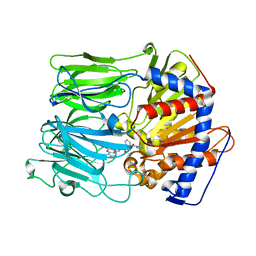 | | Prolyl oligopeptidase complexed with R-Pro-(decarboxy-Pro)-Type inhibitors | | Descriptor: | 1-{3-oxo-3-[(2S)-2-(pyrrolidin-1-ylcarbonyl)pyrrolidin-1-yl]propyl}-3-phenylquinoxalin-2(1H)-one, Prolyl endopeptidase | | Authors: | Kanai, K, Aranyi, P, Bocskei, Z, Ferenczy, G, Harmat, V, Simon, K, Naray-Szabo, G, Hermecz, I. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Prolyl oligopeptidase inhibition by N-acyl-pro-pyrrolidine-type molecules
J.Med.Chem., 51, 2008
|
|
3EQ9
 
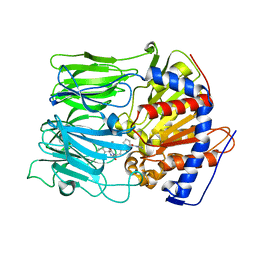 | | Prolyl oligopeptidase complexed with R-Pro-(decarboxy-Pro)-Type inhibitors | | Descriptor: | 3-{4-oxo-4-[(2S)-2-(pyrrolidin-1-ylcarbonyl)pyrrolidin-1-yl]butyl}-5,5-diphenylimidazolidine-2,4-dione, Prolyl endopeptidase | | Authors: | Kanai, K, Aranyi, P, Bocskei, Z, Ferenczy, G, Harmat, V, Simon, K, Naray-Szabo, G, Hermecz, I. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Prolyl oligopeptidase inhibition by N-acyl-pro-pyrrolidine-type molecules
J.Med.Chem., 51, 2008
|
|
4HXF
 
 | | Acylaminoacyl peptidase in complex with Z-Gly-Gly-Phe-chloromethyl ketone | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Kiss-Szeman, A, Menyhard, D.K, Tichy-Racs, E, Hornung, B, Radi, K, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
4HXE
 
 | | Pyrococcus horikoshii acylaminoacyl peptidase (uncomplexed) | | Descriptor: | HEXANE-1,6-DIOL, MAGNESIUM ION, Putative uncharacterized protein PH0594 | | Authors: | Tichy-Racs, E, Hornung, B, Radi, K, Menyhard, D.K, Kiss-Szeman, A, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
4HXG
 
 | | Pyrococcus horikoshii acylaminoacyl peptidase (orthorhombic crystal form) | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Kiss-Szeman, A, Menyhard, D.K, Tichy-Racs, E, Hornung, B, Radi, K, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
1XLM
 
 | | D254E, D256E MUTANT OF D-XYLOSE ISOMERASE COMPLEXED WITH AL3 AND XYLITOL | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Gerczei, T, Bocskei, Z.S, Szabo, E, Naray-Szabo, G, Asboth, B. | | Deposit date: | 1997-07-22 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination and refinement of the Al3+ complex of the D254,256E mutant of Arthrobacter D-xylose isomerase at 2.40 A resolution. Further evidence for inhibitor-induced metal ion movement.
Int.J.Biol.Macromol., 25, 1999
|
|
1QIW
 
 | | Calmodulin complexed with N-(3,3,-diphenylpropyl)-N'-[1-R-(3,4-bis-butoxyphenyl)-ethyl]-propylenediamine (DPD) | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
1Q3X
 
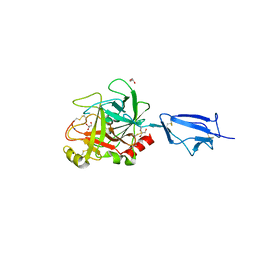 | | Crystal structure of the catalytic region of human MASP-2 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 2, SODIUM ION | | Authors: | Harmat, V, Gal, P, Kardos, J, Szilagyi, K, Ambrus, G, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions
J.Mol.Biol., 342, 2004
|
|
2QY0
 
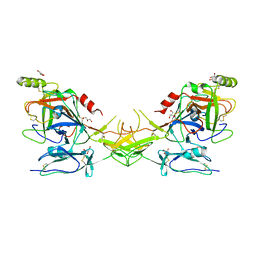 | | Active dimeric structure of the catalytic domain of C1r reveals enzyme-product like contacts | | Descriptor: | Complement C1r subcomponent, GLYCEROL | | Authors: | Kardos, J, Harmat, V, Pallo, A, Barabas, O, Szilagyi, K, Graf, L, Naray-Szabo, G, Goto, Y, Zavodszky, P, Gal, P. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Mol.Immunol., 45, 2008
|
|
1KF0
 
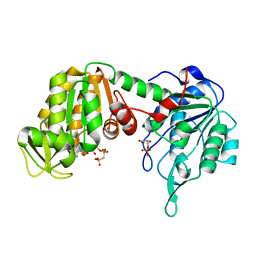 | | Crystal Structure of Pig Muscle Phosphoglycerate Kinase Ternary Complex with AMP-PCP and 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Kovari, Z, Flachner, B, Naray-Szabo, G, Vas, M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and thiol-reactivity studies on the complex of pig muscle phosphoglycerate kinase with ATP analogues: correlation between nucleotide binding mode and helix flexibility.
Biochemistry, 41, 2002
|
|
1KDQ
 
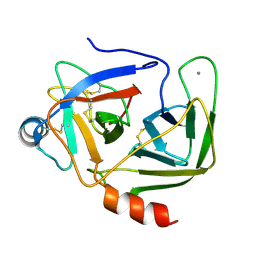 | | Crystal Structure Analysis of the Mutant S189D Rat Chymotrypsin | | Descriptor: | CALCIUM ION, CHYMOTRYPSIN B, B CHAIN, ... | | Authors: | Szabo, E, Bocskei, Z, Naray-Szabo, G, Graf, L, Venekei, I. | | Deposit date: | 2001-11-13 | | Release date: | 2003-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three Dimensional Structures of S189D Chymotrypsin and D189S Trypsin Mutants: The Effect of Polarity at Site 189 on a Protease-specific Stabilization of
the Substrate-binding Site
J.Mol.Biol., 331, 2003
|
|
3O4H
 
 | | Structure and Catalysis of Acylaminoacyl Peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, SODIUM ION | | Authors: | Harmat, V, Domokos, K, Menyhard, D.K, Pallo, A, Szeltner, Z, Szamosi, I, Beke-Somfai, T, Naray-Szabo, G, Polgar, L. | | Deposit date: | 2010-07-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure and Catalysis of Acylaminoacyl Peptidase: CLOSED AND OPEN SUBUNITS OF A DIMER OLIGOPEPTIDASE.
J.Biol.Chem., 286, 2011
|
|
3O4J
 
 | | Structure and Catalysis of Acylaminoacyl Peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Harmat, V, Domokos, K, Menyhard, D.K, Pallo, A, Szeltner, Z, Szamosi, I, Beke-Somfai, T, Naray-Szabo, G, Polgar, L. | | Deposit date: | 2010-07-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Catalysis of Acylaminoacyl Peptidase: CLOSED AND OPEN SUBUNITS OF A DIMER OLIGOPEPTIDASE.
J.Biol.Chem., 286, 2011
|
|
3O4I
 
 | | Structure and Catalysis of Acylaminoacyl Peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, CHLORIDE ION, GLYCEROL | | Authors: | Harmat, V, Domokos, K, Menyhard, D.K, Pallo, A, Szeltner, Z, Szamosi, I, Beke-Somfai, T, Naray-Szabo, G, Polgar, L. | | Deposit date: | 2010-07-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Catalysis of Acylaminoacyl Peptidase: CLOSED AND OPEN SUBUNITS OF A DIMER OLIGOPEPTIDASE.
J.Biol.Chem., 286, 2011
|
|
3O4G
 
 | | Structure and Catalysis of Acylaminoacyl Peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Harmat, V, Domokos, K, Menyhard, D.K, Pallo, A, Szeltner, Z, Szamosi, I, Beke-Somfai, T, Naray-Szabo, G, Polgar, L. | | Deposit date: | 2010-07-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Catalysis of Acylaminoacyl Peptidase: CLOSED AND OPEN SUBUNITS OF A DIMER OLIGOPEPTIDASE.
J.Biol.Chem., 286, 2011
|
|
1XA5
 
 | | Structure of Calmodulin in complex with KAR-2, a bis-indol alkaloid | | Descriptor: | 3"-(BETA-CHLOROETHYL)-2",4"-DIOXO-3, 5"-SPIRO-OXAZOLIDINO-4-DEACETOXY-VINBLASTINE, CALCIUM ION, ... | | Authors: | Horvath, I, Harmat, V, Hlavanda, E, Naray-Szabo, G, Ovadi, J. | | Deposit date: | 2004-08-25 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of the complex of calmodulin with KAR-2: a novel mode of binding explains the unique pharmacology of the drug
J.Biol.Chem., 280, 2005
|
|
2QR5
 
 | | Aeropyrum pernix acylaminoacyl peptidase, H367A mutant | | Descriptor: | Acylamino-acid-releasing enzyme | | Authors: | Harmat, V, Pallo, A, Kiss, A.L, Polgar, L. | | Deposit date: | 2007-07-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic contributions of the oxyanion binding site to the catalytic activity of acylaminoacyl peptidase
J.Struct.Biol., 162, 2008
|
|
1VJD
 
 | | Structure of pig muscle PGK complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, phosphoglycerate kinase | | Authors: | Flachner, B, Kovari, Z, Varga, A, Gugolya, Z, Vonderviszt, F, Naray-Szabo, G, Vas, M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of phosphate chain mobility of MgATP in completing the 3-phosphoglycerate kinase catalytic site: binding, kinetic, and crystallographic studies with ATP and MgATP.
Biochemistry, 43, 2004
|
|
1VJC
 
 | | Structure of pig muscle PGK complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, phosphoglycerate kinase | | Authors: | Flachner, B, Kovari, Z, Varga, A, Gugolya, Z, Vonderviszt, F, Naray-Szabo, G, Vas, M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of phosphate chain mobility of MgATP in completing the 3-phosphoglycerate kinase catalytic site: binding, kinetic, and crystallographic studies with ATP and MgATP.
Biochemistry, 43, 2004
|
|
