2L6W
 
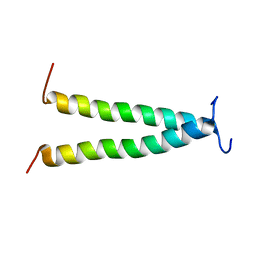 | | PDGFR beta-TM | | Descriptor: | Beta-type platelet-derived growth factor receptor | | Authors: | Muhle-Goll, C, Hoffmann, S, Ulrich, A.S. | | Deposit date: | 2010-11-29 | | Release date: | 2012-05-30 | | Last modified: | 2014-02-05 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) beta transmembrane segment.
J.Biol.Chem., 287, 2012
|
|
1XX0
 
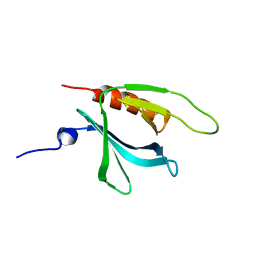 | | Structure of the C-terminal PH domain of human pleckstrin | | Descriptor: | Pleckstrin | | Authors: | Edlich, C, Stier, G, Simon, B, Sattler, M, Muhle-Goll, C. | | Deposit date: | 2004-11-03 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and phosphatidylinositol-(3,4)-bisphosphate binding of the C-terminal PH domain of human pleckstrin
STRUCTURE, 13, 2005
|
|
2A38
 
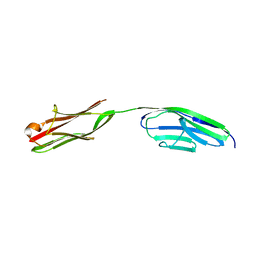 | | Crystal structure of the N-Terminus of titin | | Descriptor: | CADMIUM ION, Titin | | Authors: | Marino, M, Muhle-Goll, C, Svergun, D, Demirel, M, Mayans, O. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin
Structure, 14, 2006
|
|
7AT7
 
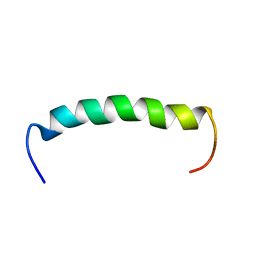 | |
7ATB
 
 | |
7ASY
 
 | |
7Z0B
 
 | |
7Z07
 
 | |
7Z08
 
 | |
7YYI
 
 | |
8ORY
 
 | |
8OS0
 
 | |
8OR5
 
 | |
8ORZ
 
 | |
7QAL
 
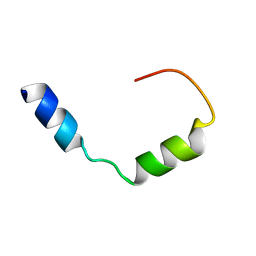 | | Three-dimensional structure of the PGAM5 C12L mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAP
 
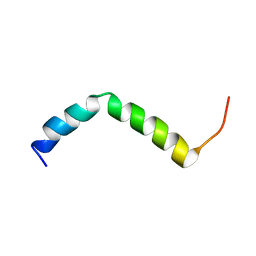 | | Three-dimensional structure of the PGAM5 G17L mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAM
 
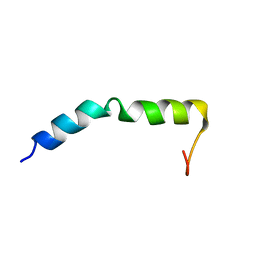 | | Three-dimensional structure of the PGAM5 WT TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAO
 
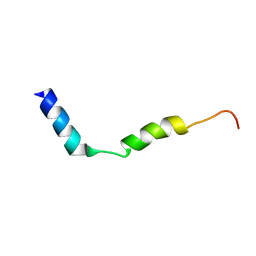 | | Three-dimensional structure of the PGAM5 C12S mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
2O10
 
 | |
2O13
 
 | |
1OG7
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-04-25 | | Release date: | 2003-09-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OHN
 
 | | Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
1OHM
 
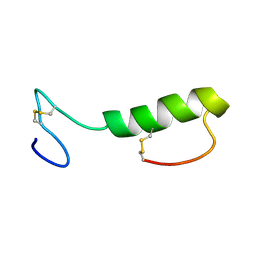 | | Sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. | | Descriptor: | BACTERIOCIN SAKACIN P | | Authors: | Uteng, M, Hauge, H.H, Markwick, P.R, Fimland, G, Mantzilas, D, Nissen-Meyer, J, Muhle-Goll, C. | | Deposit date: | 2003-05-28 | | Release date: | 2003-09-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure in Lipid Micelles of the Pediocin-Like Antimicrobial Peptide Sakacin P and a Sakacin P Variant that is Structurally Stabilized by an Inserted C-Terminal Disulfide Bridge
Biochemistry, 42, 2003
|
|
2JWP
 
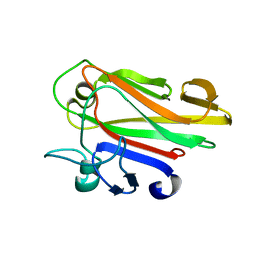 | | Malectin | | Descriptor: | Malectin | | Authors: | Schallus, T, Muhle-goll, C. | | Deposit date: | 2007-10-23 | | Release date: | 2008-08-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
6YHF
 
 | | Solution NMR Structure of APP TMD | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-29 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-23 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
