1IH5
 
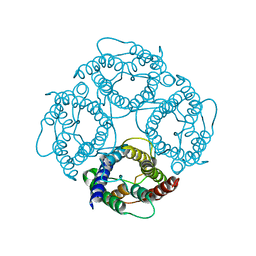 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2X8Q
 
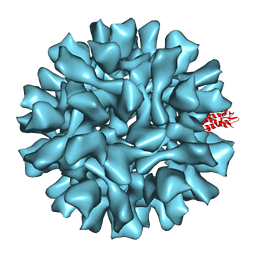 | |
5HGI
 
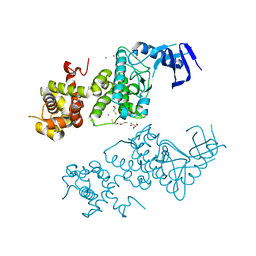 | | Crystal structure of apo human IRE1 alpha | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CESIUM ION, ... | | Authors: | Feldman, H.C, Tong, M, Wang, L, Meza-Acevedo, R, Gobillot, T.A, Gliedt, J.M, Hari, S.B, Mitra, A.K, Backes, B.J, Papa, F.R, Seeliger, M.A, Maly, D.J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Inhibition of IRE1 alpha with ATP-Competitive Ligands.
Acs Chem.Biol., 11, 2016
|
|
3TIR
 
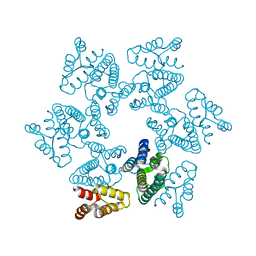 | |
3SAM
 
 | |
4E1P
 
 | |
4E1R
 
 | |
6RBK
 
 | |
6RBN
 
 | |
6RC8
 
 | |
6RAP
 
 | |
6RAO
 
 | |
6RGL
 
 | |
6SD8
 
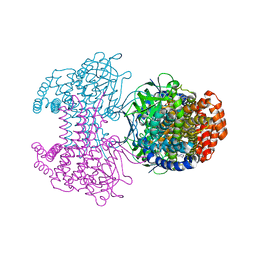 | | Bd2924 apo-form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6SDA
 
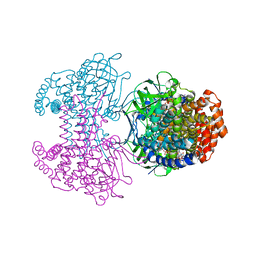 | | Bd2924 C10 acyl-coenzymeA bound form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable acyl-CoA dehydrogenase, decanoyl-CoA | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-11 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
6CVZ
 
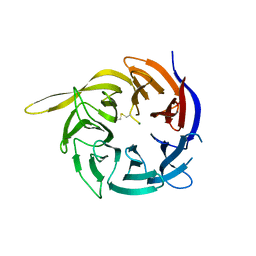 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
3G0V
 
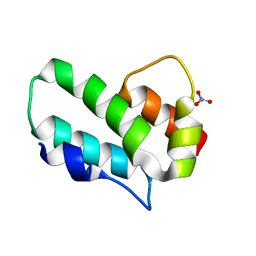 | |
3G21
 
 | |
3G28
 
 | |
3G1G
 
 | |
3G29
 
 | |
3G1I
 
 | |
3G26
 
 | |
5JCG
 
 | |
5J5S
 
 | | Src kinase in complex with a sulfonamide inhibitor | | Descriptor: | N-{4-[8-amino-3-(propan-2-yl)imidazo[1,5-a]pyrazin-1-yl]naphthalen-1-yl}-N'-[3-(trifluoromethyl)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lebedev, I, Feldman, H.C, Georghiou, G, Maly, D.J, Seeliger, M. | | Deposit date: | 2016-04-03 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Modulation of IRE1a with ATP-Competitive Kinase Inhibitors
To Be Published
|
|
