5PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (ATRAZINE COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ISOPROPYLAMINO-6-ETHYLAMINO -1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
6PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420314 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(S(-)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-31 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
7PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420315 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(R(+)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-08-01 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
7NB6
 
 | |
2PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (UBIQUINONE-2 COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
3PRC
 
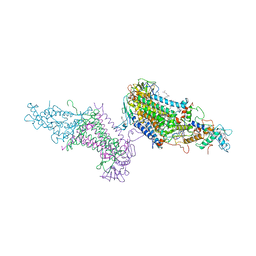 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
6YUZ
 
 | | Homodimeric structure of the rBAT complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YUP
 
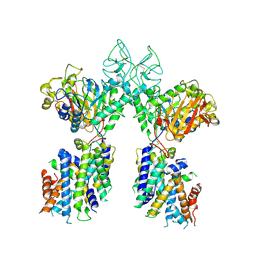 | | Heterotetrameric structure of the rBAT-b(0,+)AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT, ... | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YV1
 
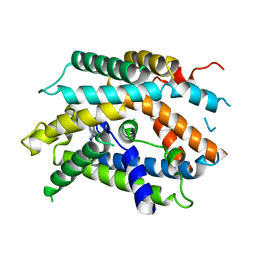 | | Structure of human b(0,+)AT1 | | Descriptor: | b(0,+)-type amino acid transporter 1 | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z71
 
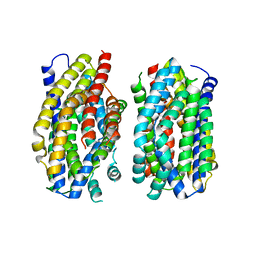 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
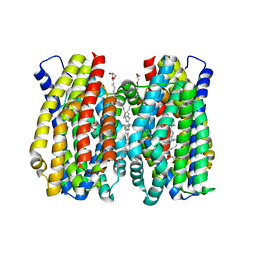 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKZ
 
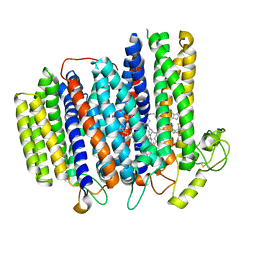 | | Cryo-EM structure of the cytochrome bd oxidase from M. tuberculosis at 2.5 A resolution | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, HEME B/C, MENAQUINONE-9, ... | | Authors: | Safarian, S, Wu, D, Krause, K.L, Michel, H. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The cryo-EM structure of the bd oxidase from M. tuberculosis reveals a unique structural framework and enables rational drug design to combat TB.
Nat Commun, 12, 2021
|
|
1DXR
 
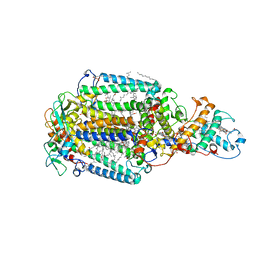 | | Photosynthetic reaction center from Rhodopseudomonas viridis - His L168 Phe mutant (terbutryn complex) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Bibikova, M, Sabatino, P, Oesterhelt, D, Michel, H. | | Deposit date: | 2000-01-15 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the drastically increased initial electron transfer rate in the reaction center from a Rhodopseudomonas viridis mutant described at 2.00-A resolution.
J. Biol. Chem., 275, 2000
|
|
1MQK
 
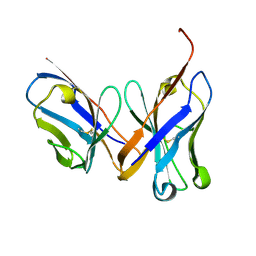 | | Crystal structure of the unliganded Fv-fragment of the anti-cytochrome C oxidase antibody 7E2 | | Descriptor: | antibody 7E2 FV fragment, heavy chain, light chain | | Authors: | Essen, L.-O, Harrenga, A, Ostermeier, C, Michel, H. | | Deposit date: | 2002-09-16 | | Release date: | 2003-04-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.3 A X-ray structure of an antibody Fv fragment used for induced membrane-protein crystallization.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PRC
 
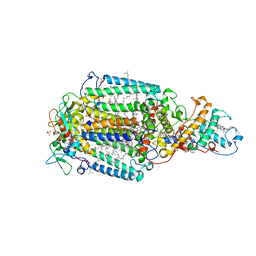 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
5IR6
 
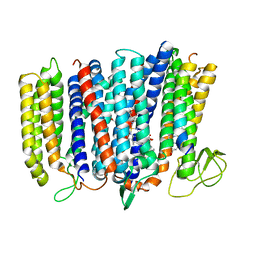 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | Descriptor: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | Deposit date: | 2016-03-12 | | Release date: | 2016-05-04 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
5DJQ
 
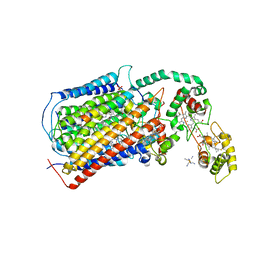 | | The structure of CBB3 cytochrome oxidase. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | Authors: | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | Deposit date: | 2015-09-02 | | Release date: | 2016-01-13 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
7OY2
 
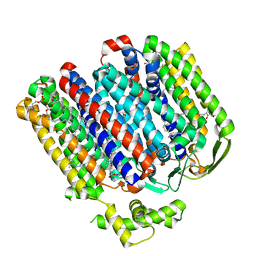 | | High resolution structure of cytochrome bd-II oxidase from E. coli | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(2~{E},6~{E},10~{Z},14~{E},18~{E},22~{E},26~{E})-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaenyl]naphthalene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Grund, T.N, Wu, D, Bald, D, Michel, H, Safarian, S. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Mechanistic and structural diversity between cytochrome bd isoforms of Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OT9
 
 | |
4MLB
 
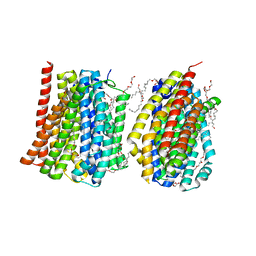 | | Reverse polarity of binding pocket suggests different function of a MOP superfamily transporter from Pyrococcus furiosus Vc1 (DSM3638) | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, PF0708 | | Authors: | Malviya, V.N, Nonaka, T, Muenke, C, Koepke, J, Michel, H. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1W2L
 
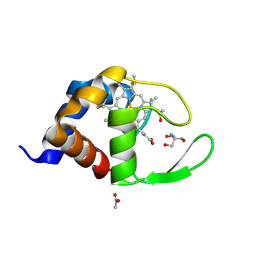 | | Cytochrome c domain of caa3 oxygen oxidoreductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CYTOCHROME OXIDASE SUBUNIT II, ... | | Authors: | Srinivasan, V, Rajendran, C, Sousa, F.L, Melo, A.M.P, Saraiva, L.M, Pereira, M.M, Santana, M, Teixeira, M, Michel, H. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure at 1.3 A resolution of Rhodothermus marinus caa(3) cytochrome c domain.
J. Mol. Biol., 345, 2005
|
|
1ZCD
 
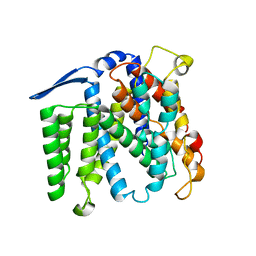 | | Crystal structure of the Na+/H+ antiporter NhaA | | Descriptor: | Na(+)/H(+) antiporter 1 | | Authors: | Hunte, C, Screpanti, E, Venturi, M, Rimon, A, Padan, E, Michel, H. | | Deposit date: | 2005-04-11 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH.
Nature, 435, 2005
|
|
1F6N
 
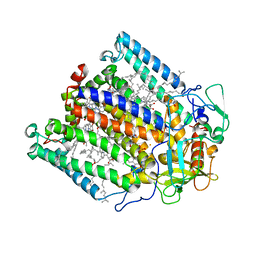 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> TYR FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-06-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
1FNQ
 
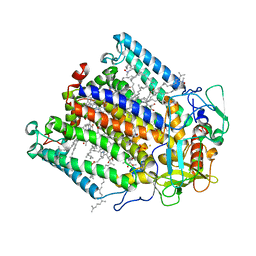 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> GLU FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
1FNP
 
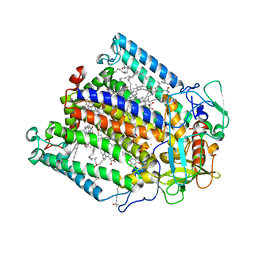 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> PHE FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
