7AVR
 
 | | The tetrameric structure of haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase 1 | | Authors: | Mazur, A, Kolenko, P, Prudnikova, T, Grinkevich, P, Kuta Smatanova, I. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetrameric structure of the novel haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7PW1
 
 | | Crystal structure of ancestral haloalkane dehalogenase AncLinB-DmbA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Mazur, A, Grinkevich, P, Prudnikova, T. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Ancestral Haloalkane Dehalogenase AncLinB-DmbA.
Int J Mol Sci, 22, 2021
|
|
4QPF
 
 | | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-fluoro-2-(imidazo[1,2-a]pyridin-3-yl)ethane-1,1-diyl]bis(phosphonic acid) | | Authors: | Ebetino, F.H, Lundy, M, Kwaasi, A.A, Dunford, J.E, Duan, Z, Triffitt, J, Mazur, A, Jeans, G, Barnett, B.L, Russell, R.G.G. | | Deposit date: | 2014-06-23 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | New lower bone affinity bisphosphonate drug design for effective use in diseases characterized by abnormal bone resorption
To be Published
|
|
4C4Z
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Y
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A4 | | Descriptor: | 1-(3-chlorophenyl)-3-(2-methoxyethyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4X
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4IE1
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 1h | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-8-hydroxyoctyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IE3
 
 | | Crystal structure of human Arginase-2 complexed with inhbitor 1o | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3522 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IE2
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 1h | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2082 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2N1F
 
 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6YTC
 
 | |
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5OWI
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 1) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
6V5D
 
 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
4YXS
 
 | | CAMP-DEPENDENT PROTEIN KINASE PKA CATALYTIC SUBUNIT WITH PKI-5-24 | | Descriptor: | N-BENZYL-9H-PURIN-6-AMINE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-20 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YXR
 
 | | CRYSTAL STRUCTURE OF PKA IN COMPLEX WITH inhibitor. | | Descriptor: | 3-methyl-2H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CMT
 
 | |
5CGL
 
 | |
5CKL
 
 | |
6Z9A
 
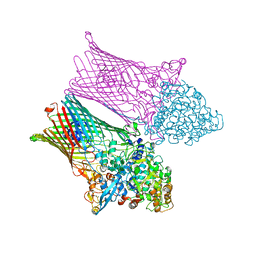 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6Z8I
 
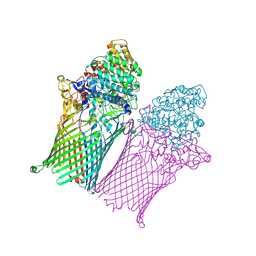 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZAZ
 
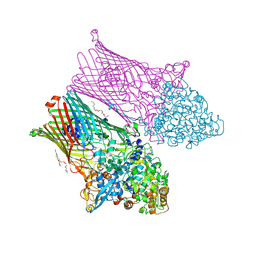 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZLU
 
 | |
6ZM1
 
 | |
