3NEZ
 
 | | mRojoA | | Descriptor: | mRojoA | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NF0
 
 | | mPlum-TTN | | Descriptor: | D-MALATE, Fluorescent protein plum | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Changes in the Fluorescent Properties of Computationally Designed Red Fluorescent Proteins
To be Published
|
|
3NED
 
 | | mRouge | | Descriptor: | ACETATE ION, PAmCherry1 protein, SODIUM ION | | Authors: | Mayo, S.L, Chica, R.A, Moore, M.M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
4PT6
 
 | | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities | | Descriptor: | Coagulation factor VIII | | Authors: | Kym, G, Shi, K, Kamajaya, A, Hamdouche, S, Kostecki, J.S, Wannier, T, Li, B, Nikolovski, P, Mayo, S.L. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities
To be Published
|
|
4QTR
 
 | | Computational design of co-assembling protein-DNA nanowires | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*TP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*AP*TP*TP*TP*AP*AP*TP*TP*TP*CP*C)-3'), dualENH | | Authors: | Mou, Y, Mayo, S.L. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of co-assembling protein-DNA nanowires.
Nature, 525, 2015
|
|
4ZN8
 
 | |
1GB4
 
 | |
3QR3
 
 | | Crystal Structure of Cel5A (EG2) from Hypocrea jecorina (Trichoderma reesei) | | Descriptor: | Endoglucanase EG-II, MAGNESIUM ION, SULFATE ION | | Authors: | Lee, T.M, Farrow, M.F, Kaiser, J.T, Arnold, F.H, Mayo, S.L. | | Deposit date: | 2011-02-16 | | Release date: | 2011-11-02 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural study of Hypocrea jecorina Cel5A.
Protein Sci., 20, 2011
|
|
1Y66
 
 | | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, CADMIUM ION, ... | | Authors: | Hom, G.K, Lassila, J.K, Thomas, L.M, Mayo, S.L. | | Deposit date: | 2004-12-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant.
Protein Sci., 14, 2005
|
|
3S3Z
 
 | |
3S3Y
 
 | |
2MG4
 
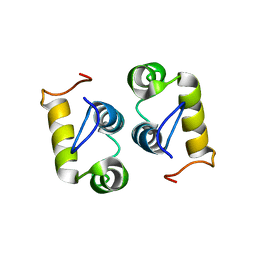 | | Computational design and experimental verification of a symmetric protein homodimer | | Descriptor: | Computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2P6J
 
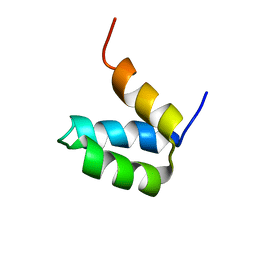 | | Full-sequence computational design and solution structure of a thermostable protein variant | | Descriptor: | designed engrailed homeodomain variant UVF | | Authors: | Shah, P.S, Hom, G.K, Ross, S.A, Lassila, J.K, Crowhurst, K.A, Mayo, S.L. | | Deposit date: | 2007-03-18 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Full-sequence Computational Design and Solution Structure of a Thermostable Protein Variant.
J.Mol.Biol., 372, 2007
|
|
4NDK
 
 | |
4NDJ
 
 | |
4NDL
 
 | | Computational design and experimental verification of a symmetric homodimer | | Descriptor: | ENH-c2b, computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OHS
 
 | | The structure of a far-red fluorescent protein, AQ143 | | Descriptor: | CHLORIDE ION, FAR-RED FLUORESCENT PROTEIN AQ143 | | Authors: | Wannier, T.M, Mayo, S.L. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-26 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of a far-red fluorescent protein, AQ143, shows evidence in support of reported red-shifting chromophore interactions.
Protein Sci., 23, 2014
|
|
6DEJ
 
 | |
3O2L
 
 | |
3NYZ
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | Descriptor: | Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NYD
 
 | | Crystal Structure of Kemp Eliminase HG-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-14 | | Release date: | 2011-06-29 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NZ1
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1PSV
 
 | |
1QCV
 
 | |
1EM7
 
 | |
