1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
1LTR
 
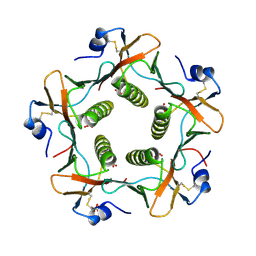 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HUMAN HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SULFATE ION | | Authors: | Matkovic-Calogovic, D, Loreggian, A, Palu, G, Zanotti, G. | | Deposit date: | 1998-07-31 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
4RXW
 
 | |
2EZ1
 
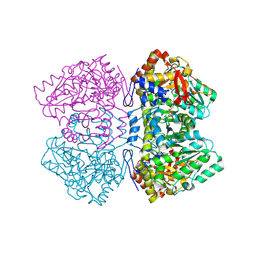 | | Holo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2EZ2
 
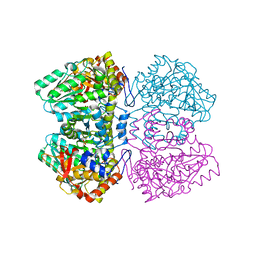 | | Apo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
5K5Y
 
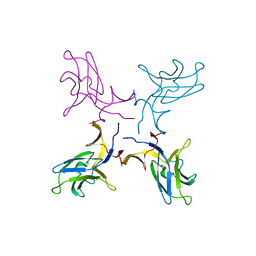 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori (strain 26695) | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Kekez, I, Cendron, L, Stojanovic, M, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Stability of FlgD from the Pathogenic 26695 Strain of Helicobacter pylori
Croatica Chemica Acta, 2016
|
|
6GIR
 
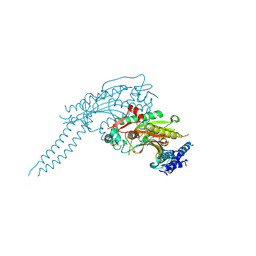 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
2VLH
 
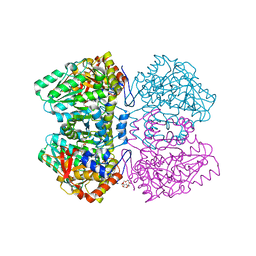 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VLF
 
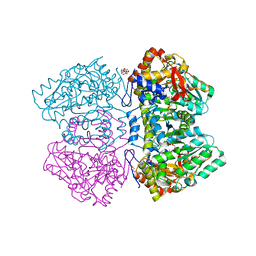 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
5NPY
 
 | | Crystal structure of Helicobacter pylori flagellar hook protein FlgE2 | | Descriptor: | Flagellar basal body protein, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Loconte, V, Zanotti, G, Kekez, I, Matkovic-Calogovic, D. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural characterization of FlgE2 protein from Helicobacter pylori hook.
FEBS J., 284, 2017
|
|
6TG8
 
 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
3TT8
 
 | |
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
4FKA
 
 | | High resolution structure of the manganese derivative of insulin | | Descriptor: | Insulin A chain, Insulin B chain, MANGANESE (II) ION, ... | | Authors: | Prugovecki, B, Pulic, I, Toth, M, Matkovic-Calogovic, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High Resolution Structure of the Manganese Derivative of Insulin
Croat.Chem.Acta, 85, 2012
|
|
2YCT
 
 | | Tyrosine phenol-lyase from Citrobacter freundii in complex with pyridine N-oxide and the quinonoid intermediate formed with L-alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PHOSPHATE ION, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2YCP
 
 | | F448H mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2YCN
 
 | | Y71F mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2YHK
 
 | | D214A mutant of tyrosine phenol-lyase from Citrobacter freundii | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Citrobacter Freundii Asp214Ala Tyrosine Phenol-Lyase Reveals that Asp214 is Critical for Maintaining a Strain in the Internal Aldimine
Croatica Chemica Acta, 85, 2012
|
|
4ZZF
 
 | | Crystal structure of truncated FlgD (tetragonal form) from the human pathogen Helicobacter pylori | | Descriptor: | Flagellar basal body rod modification protein | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.1673 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
4ZZK
 
 | | Crystal structure of truncated FlgD (monoclinic form) from the human pathogen Helicobacter pylori | | Descriptor: | Basal-body rod modification protein FlgD | | Authors: | Pulic, I, Cendron, L, Salamina, M, Matkovic-Calogovic, D, Zanotti, G. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of truncated FlgD from the human pathogen Helicobacter pylori.
J.Struct.Biol., 194, 2016
|
|
3EXX
 
 | |
