5L9L
 
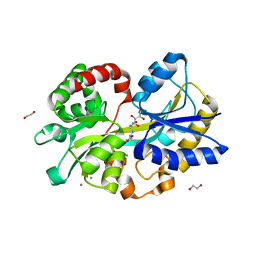 | |
5L9O
 
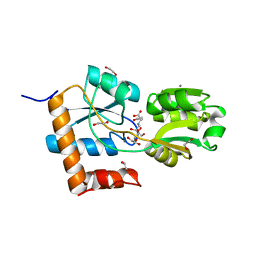 | |
5L9S
 
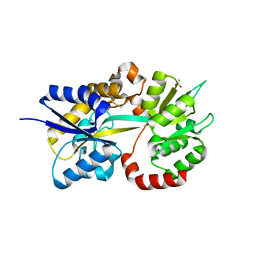 | |
5L9G
 
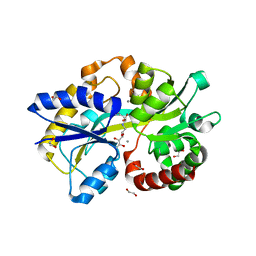 | |
5L9I
 
 | |
5LOM
 
 | | Crystal structure of the PBP SocA from Agrobacterium tumefaciens C58 in complex with DFG at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Deoxyfructosyl-amino Acid Transporter Periplasmic Binding Protein, Deoxyfructosylglutamine | | Authors: | Marty, L, Vigouroux, A, Morera, S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLZ
 
 | | Structure in C2 form of the PBP AgtB from A.tumefacien R10 in complex with agropinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Agropine permease, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HM2
 
 | | Structure in P1 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, SODIUM ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HLX
 
 | | Structure of the PBP AgaA in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
6HQH
 
 | |
5L9P
 
 | | Crystal structure of the PBP MotA from A. tumefaciens B6 | | Descriptor: | SULFATE ION, periplasmic binding protein | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2016-06-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
6R3Z
 
 | |
6R44
 
 | |
6TG2
 
 | | Structure of the PBP/SBP MotA in complex with mannopinic acid from A.tumefacien R10 | | Descriptor: | (2~{R})-2-[[(3~{R},4~{R},5~{S})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S, Marty, L. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
5NV4
 
 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum double mutant D611C:G1050C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5F0E
 
 | | Murine endoplasmic reticulum alpha-glucosidase II | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-11-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.106 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6R5S
 
 | |
6R6K
 
 | | Structure of a FpvC mutant from pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa.
Febs J., 287, 2020
|
|
6RU4
 
 | |
6TG3
 
 | | Crystal Structure of the PBP/SBP MotA in complex with glucopinic acid from A. tumefaciens B6/R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, MotA | | Authors: | Morera, S, Vigouroux, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
