1ZMR
 
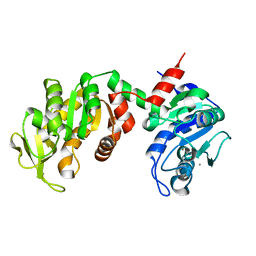 | | Crystal Structure of the E. coli Phosphoglycerate Kinase | | Descriptor: | CALCIUM ION, Phosphoglycerate kinase | | Authors: | Marqusee, S. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of proteolytic susceptibility in phosphoglycerate kinases from yeast and E. coli: modulation of conformational ensembles without altering structure or stability.
J.Mol.Biol., 368, 2007
|
|
1DOF
 
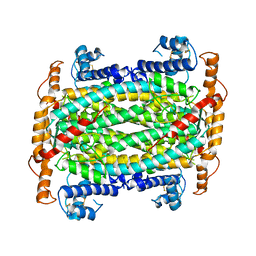 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
3H08
 
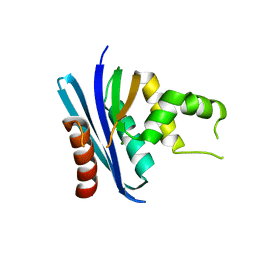 | | Crystal structure of the Ribonuclease H1 from Chlorobium tepidum | | Descriptor: | MAGNESIUM ION, Rnh (Ribonuclease H) | | Authors: | Ratcliff, K, Corn, J, Marqusee, S. | | Deposit date: | 2009-04-08 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, stability, and folding of ribonuclease H1 from the moderately thermophilic chlorobium tepidum: comparison with thermophilic and mesophilic homologues.
Biochemistry, 48, 2009
|
|
8TQO
 
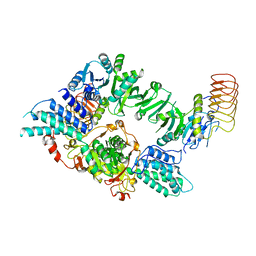 | | Eukaryotic translation initiation factor 2B tetramer | | Descriptor: | Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, Translation initiation factor eIF-2B subunit epsilon, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
8TQZ
 
 | | Eukaryotic translation initiation factor 2B with a mutation (L516A) in the delta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
4LY7
 
 | | Ancestral RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Hart, K.M, Harms, M.J, Schmidt, B.H, Elya, C, Thornton, J.W, Marqusee, S. | | Deposit date: | 2013-07-30 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Thermodynamic system drift in protein evolution.
Plos Biol., 12, 2014
|
|
7UI5
 
 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1JXB
 
 | |
1JL2
 
 | |
1F21
 
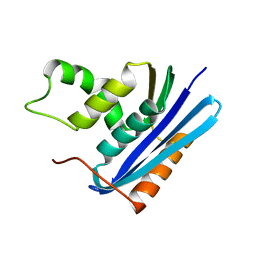 | |
1G15
 
 | |
2II0
 
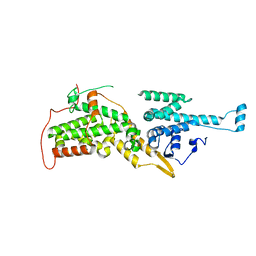 | | Crystal Structure of catalytic domain of Son of sevenless (Rem-Cdc25) in the absence of Ras | | Descriptor: | Son of sevenless homolog 1 | | Authors: | Freedman, T.S, Sondermann, H, Friedland, G.D, Kortemme, T, Bar-Sagi, D, Marqusee, S, Kuriyan, J. | | Deposit date: | 2006-09-27 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Ras-induced conformational switch in the Ras activator Son of sevenless.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1JL1
 
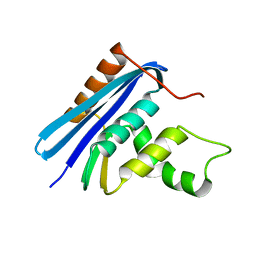 | | D10A E. coli ribonuclease HI | | Descriptor: | RIBONUCLEASE HI | | Authors: | Goedken, E.R, Marqusee, S. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Native-state energetics of a thermostabilized variant of ribonuclease HI.
J.Mol.Biol., 314, 2001
|
|
2O79
 
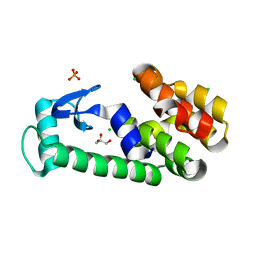 | | T4 lysozyme with C-terminal extension | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme, ... | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
2O4W
 
 | | T4 lysozyme circular permutant | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
6XHZ
 
 | | Alpha-lytic protease homolog N4 | | Descriptor: | N4: hypothetical protein, SULFATE ION | | Authors: | Nixon, C.F, Marqusee, S.M, Gee, C.L. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploring the Evolutionary History of Kinetic Stability in the alpha-Lytic Protease Family.
Biochemistry, 60, 2021
|
|
6UEW
 
 | |
7SA7
 
 | |
8FU6
 
 | | GCGR-Gs complex in the presence of RAMP2 | | Descriptor: | Glucagon derivative ZP3780, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, O'Brien, E.S, Wang, H, Montabana, E, Kobilka, B.K. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Negative allosteric modulation of the glucagon receptor by RAMP2.
Cell, 186, 2023
|
|
2O7A
 
 | | T4 lysozyme C-terminal fragment | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme | | Authors: | Echols, N, Kwon, E, Marqusee, S.M, Alber, T. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
4Z0U
 
 | | RNase HI/SSB-Ct complex | | Descriptor: | Ribonuclease H, SSB-Ct Peptide | | Authors: | Petzold, C, Keck, J.L. | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity.
J.Biol.Chem., 290, 2015
|
|
1F1O
 
 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
2IJE
 
 | |
8T8M
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Step-wise activation of a Family C GPCR.
Biorxiv, 2023
|
|
8T6J
 
 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Step-wise activation of a Family C GPCR.
Biorxiv, 2023
|
|
