1D6J
 
 | |
1I2D
 
 | |
1M8P
 
 | |
2QVW
 
 | |
2FFL
 
 | |
7SWF
 
 | |
7SVA
 
 | | Cryo-EM structure of Arabidopsis Ago10-guide RNA complex | | Descriptor: | MAGNESIUM ION, Protein argonaute 10, RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP*UP*UP*U)-3') | | Authors: | Xiao, Y, MacRae, I.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for RNA slicing by a plant Argonaute.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7SWQ
 
 | |
7KI3
 
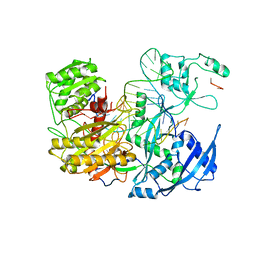 | |
5WEA
 
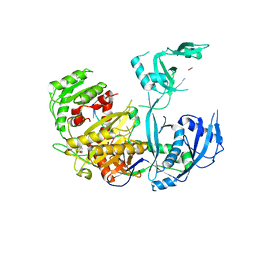 | | Human Argonaute2 Helix-7 Mutant | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*U)-3') | | Authors: | Schirle, N.T, Klum, S.M, MacRae, I.J. | | Deposit date: | 2017-07-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition.
EMBO J., 37, 2018
|
|
7KX7
 
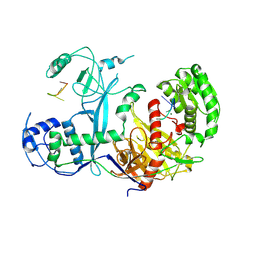 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*AP*GP*(OMC))-3') | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
7KX9
 
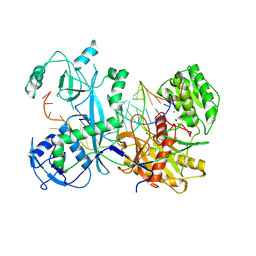 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA-target complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*UP*UP*GP*AP*GP*UP*UP*GP*GP*AP*CP*AP*AP*AP*UP*GP*GP*CP*AP*(OMG))-3'), ... | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
6NIT
 
 | |
3KS4
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
4GHA
 
 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
4GH9
 
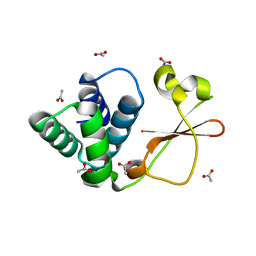 | | Crystal structure of Marburg virus VP35 RNA binding domain | | Descriptor: | ACETATE ION, Polymerase cofactor VP35 | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
3Q7C
 
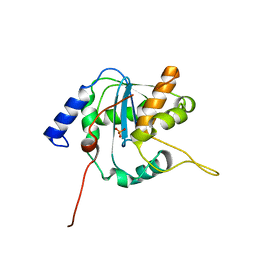 | | Exonuclease domain of Lassa virus nucleoprotein bound to manganese | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, PHOSPHATE ION, ... | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q7B
 
 | | Exonuclease domain of Lassa virus nucleoprotein | | Descriptor: | Nucleoprotein, PHOSPHATE ION, ZINC ION | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4OLB
 
 | |
4OLA
 
 | | Crystal Structure of Human Argonaute2 | | Descriptor: | 5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*U)-3', ISOPROPYL ALCOHOL, PHENOL, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human Argonaute2.
Science, 336, 2012
|
|
4W5R
 
 | |
4W5Q
 
 | |
4W5T
 
 | |
4W5O
 
 | |
