3LH5
 
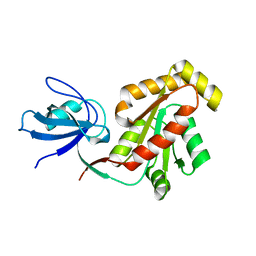 | |
5DOE
 
 | | Crystal structure of the Human Cytomegalovirus UL53 (residues 72-292) | | Descriptor: | Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
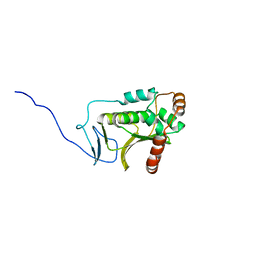
5DOB
 
 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
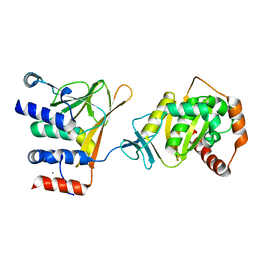
5DOC
 
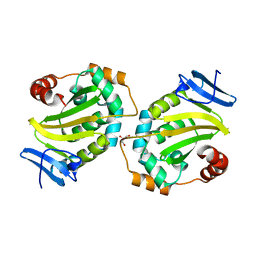 | | Crystal structure of the Human Cytomegalovirus UL53 subunit of the NEC | | Descriptor: | GLYCEROL, Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
2RCZ
 
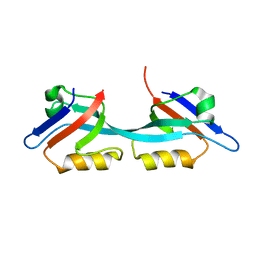 | | Structure of the second PDZ domain of ZO-1 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Lavie, A, Lye, M.F. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Domain swapping within PDZ2 is responsible for dimerization of ZO proteins.
J.Biol.Chem., 282, 2007
|
|
3TSZ
 
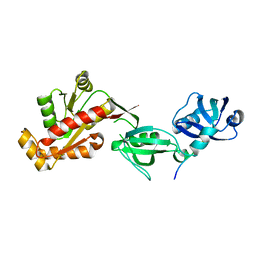 | |
3TSW
 
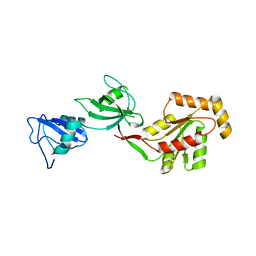 | | crystal structure of the PDZ3-SH3-GUK core module of Human ZO-1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The Src Homology 3 Domain Is Required for Junctional Adhesion Molecule Binding to the Third PDZ Domain of the Scaffolding Protein ZO-1.
J.Biol.Chem., 286, 2011
|
|
3TSV
 
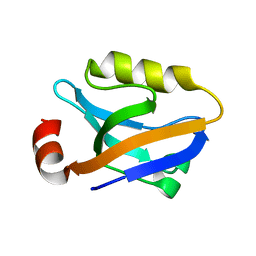 | |
