4S3H
 
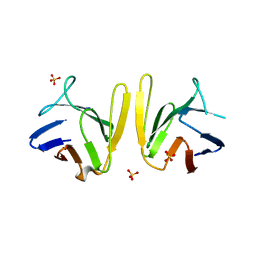 | | Crystal structure of S. pombe Mdb1 FHA domain | | Descriptor: | Mdb1, SULFATE ION | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1.
J.Biol.Chem., 290, 2015
|
|
4U8T
 
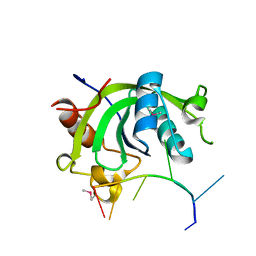 | |
6E40
 
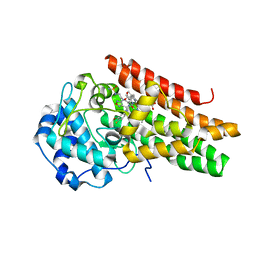 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complexed with ferric heme and Epacadostat | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E42
 
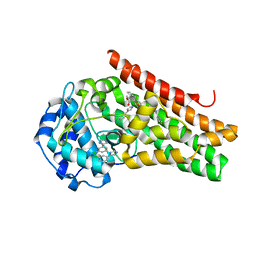 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and 4-Chlorophenyl imidazole | | Descriptor: | 4-(3-chlorophenyl)-1H-imidazole, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E41
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and an Epacadostat analog | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]sulfanyl}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E43
 
 | | Crystal structure of human indoleamine 2,3-dioxygenase 1 (IDO1) in complex with a BMS-978587 analog | | Descriptor: | (1R,2S)-2-(4-[cyclohexyl(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, BENZOIC ACID, Indoleamine 2,3-dioxygenase 1 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E45
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferrous state | | Descriptor: | GLYCEROL, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E44
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferric state | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E46
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferrous heme and tryptophan | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3UV0
 
 | | Crystal structure of the drosophila MU2 FHA domain | | Descriptor: | Mutator 2, isoform B | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimerization, but not phosphothreonine binding, is conserved between the forkhead-associated domains of Drosophila MU2 and human MDC1
Febs Lett., 586, 2012
|
|
7KZ8
 
 | |
7KZ9
 
 | | Crystal structure of Pseudomonas sp. PDC86 substrate-binding protein Aapf in complex with a signaling molecule HEHEAA | | Descriptor: | GLYCEROL, N,N~2~-bis(2-hydroxyethyl)glycinamide, Peptide/nickel transport system substrate-binding protein AapF, ... | | Authors: | Luo, S, Dadhwal, P, Tong, L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a bacterial Pip system plant effector recognition protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5U1T
 
 | |
5U1S
 
 | |
6JIW
 
 | | Crystal structure of S. aureus CntK with C72S mutation | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
6JIS
 
 | | Crystal structure of the histidine racemase CntK in cobalt and nickel transporter system of staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
2M3I
 
 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | Descriptor: | Alpha-conotoxin | | Authors: | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | Deposit date: | 2013-01-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
2LZ5
 
 | | Solution structure of a Novel Alpha-Conotoxin TxIB | | Descriptor: | Conotoxin_TxIB | | Authors: | Luo, S, Zhangsun, D, Wu, Y, Zhu, X, Hu, Y, McIntyre, M, Christensen, S, Akcan, M, Craik, D, McIntosh, J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors.
J.Biol.Chem., 288, 2013
|
|
4S1C
 
 | |
4S1B
 
 | | Crystal Structure of L. monocytogenes phosphodiesterase PgpH HD domain in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, FE (III) ION, Lmo1466 protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6XNU
 
 | |
6XNV
 
 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES CBPB PROTEIN (LMO1009) IN COMPLEX WITH C-DI-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (p)ppGpp and c-di-AMP Homeostasis Is Controlled by CbpB in Listeria monocytogenes.
Mbio, 11, 2020
|
|
4JDE
 
 | | Crystal structure of PUD-1/PUD-2 heterodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein F15E11.1, ... | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2013-02-25 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of PUD-1 and PUD-2, two proteins up-regulated in a long-lived daf-2 mutant.
Plos One, 8, 2013
|
|
7UPX
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
