3U1M
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3U1L
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
3CBB
 
 | | Crystal Structure of Hepatocyte Nuclear Factor 4alpha in complex with DNA: Diabetes Gene Product | | Descriptor: | Hepatocyte Nuclear Factor 4-alpha promoter element DNA, Hepatocyte Nuclear Factor 4-alpha, DNA binding domain, ... | | Authors: | Lu, P, Rha, G.B, Melikishvili, M, Adkins, B.C, Fried, M.G, Chi, Y.I. | | Deposit date: | 2008-02-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4alpha. Diabetes gene product.
J.Biol.Chem., 283, 2008
|
|
2H8R
 
 | | Hepatocyte Nuclear Factor 1b bound to DNA: MODY5 Gene Product | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*G)-3', 5'-D(*GP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3', Hepatocyte nuclear factor 1-beta | | Authors: | Lu, P, Rha, G.B, Chi, Y.I. | | Deposit date: | 2006-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of disease-causing mutations in hepatocyte nuclear factor 1beta.
Biochemistry, 46, 2007
|
|
4O79
 
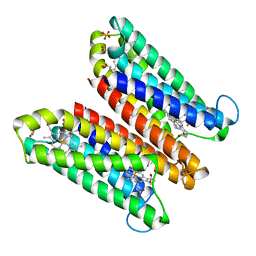 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 10 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O6Y
 
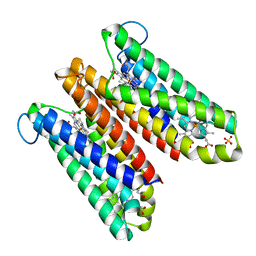 | | Crystal Structure of Cytochrome b561 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, SULFATE ION | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O7G
 
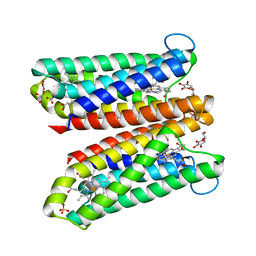 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 40 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8J4H
 
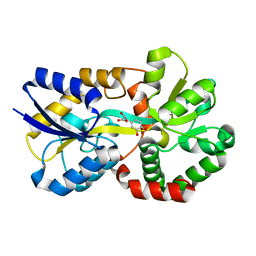 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with Danshensu (DSS) | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-propanoic acid, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8J4J
 
 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with ferric ion | | Descriptor: | CARBONATE ION, FE (III) ION, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
6B85
 
 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
7W3W
 
 | | X-ray structure of apo-VmFbpA, a ferric ion-binding protein from Vibrio metschnikovii | | Descriptor: | Iron-utilization periplasmic protein | | Authors: | Lu, P, Sui, M, Zhang, M, Nagata, K. | | Deposit date: | 2021-11-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Rosmarinic Acid and Sodium Citrate Have a Synergistic Bacteriostatic Effect against Vibrio Species by Inhibiting Iron Uptake.
Int J Mol Sci, 22, 2021
|
|
7YEH
 
 | | Cryo-EM structure of human OGT-OGA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Lu, P, Liu, Y, Yu, H, Gao, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
7YEA
 
 | | Human O-GlcNAc transferase Dimer | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gao, H, Lu, P, Liu, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
6U1S
 
 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
1FZX
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1G14
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
4R12
 
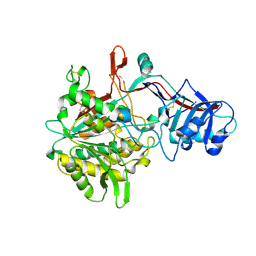 | | Crystal structure of the gamma-secretase component Nicastrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | Deposit date: | 2014-08-03 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1LBG
 
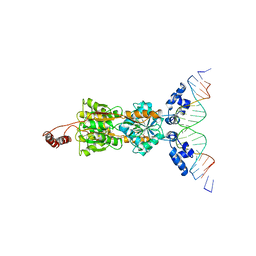 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBH
 
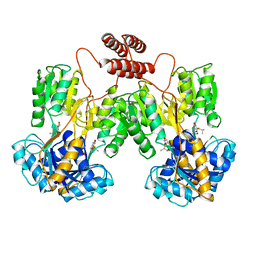 | | INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBI
 
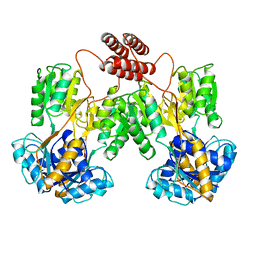 | | LAC REPRESSOR | | Descriptor: | LAC REPRESSOR | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
5A63
 
 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
8HYL
 
 | | Crystal structure of DO1 Fv-clasp fragment | | Descriptor: | VH-SARAH, VL-SARAH | | Authors: | Anan, Y, Lu, P, Nagata, K, Itakura, M, Uchida, K. | | Deposit date: | 2023-01-06 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural basis of anti-DNA antibody specificity for pyrrolated proteins
Commun Biol, 7, 2024
|
|
