1OMW
 
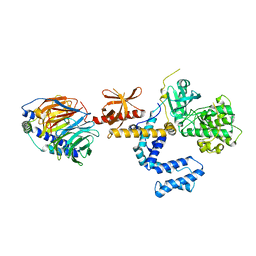 | | Crystal Structure of the complex between G Protein-Coupled Receptor Kinase 2 and Heterotrimeric G Protein beta 1 and gamma 2 subunits | | Descriptor: | G-protein coupled receptor kinase 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1 | | Authors: | Lodowski, D.T, Pitcher, J.A, Capel, W.D, Lefkowitz, R.J, Tesmer, J.J.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Keeping G proteins at Bay: A Complex Between G Protein-Coupled Receptor Kinase 2 and G-Beta-Gamma
Science, 300, 2003
|
|
2ACX
 
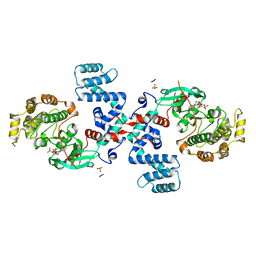 | | Crystal Structure of G protein coupled receptor kinase 6 bound to AMPPNP | | Descriptor: | G protein-coupled receptor kinase 6, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lodowski, D.T, Tesmer, V.M, Benovic, J.L, Tesmer, J.J. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of G Protein-coupled Receptor Kinase (GRK)-6 Defines a Second Lineage of GRKs.
J.Biol.Chem., 281, 2006
|
|
1YM7
 
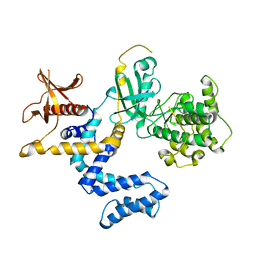 | | G Protein-Coupled Receptor Kinase 2 (GRK2) | | Descriptor: | Beta-adrenergic receptor kinase 1 | | Authors: | Lodowski, D.T, Barnhill, J.F, Pyskadlo, R.M, Ghirlando, R, Sterne-Marr, R, Tesmer, J.J.G. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The role of Gbetagamma and domain interfaces in the activation of G protein-coupled receptor kinase 2
Biochemistry, 44, 2005
|
|
2I37
 
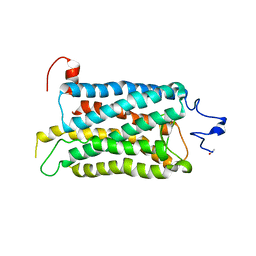 | | Crystal structure of a photoactivated rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rhodopsin, ... | | Authors: | Lodowski, D.T, Stenkamp, R.E, Salom, D, Le Trong, I, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4X1H
 
 | | Opsin/G(alpha) peptide complex stabilized by nonyl-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal derived peptide of guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Blankenship, E, Lodowski, D.T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation.
Structure, 23, 2015
|
|
2NWC
 
 | |
3KVC
 
 | | Crystal structure of bovine RPE65 at 1.9 angstrom resolution | | Descriptor: | FE (II) ION, Retinoid isomerohydrolase | | Authors: | Kiser, P.D, Lodowski, D.T, Golczak, M, Palczewski, K. | | Deposit date: | 2009-11-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of membrane structural integrity for RPE65 retinoid isomerization activity.
J.Biol.Chem., 285, 2010
|
|
4M7G
 
 | | Streptomyces Erythraeus Trypsin | | Descriptor: | Trypsin-like protease | | Authors: | Blankenship, E, Vukoti, K, Miyagi, M, Lodowski, D.T. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Conformational flexibility in the catalytic triad revealed by the high-resolution crystal structure of Streptomyces erythraeus trypsin in an unliganded state.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3CIK
 
 | | Human GRK2 in Complex with Gbetagamma subunits | | Descriptor: | Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tesmer, J.J.G, Lodowski, D.T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-02-17 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of human G protein-coupled receptor kinase 2 in complex with the kinase inhibitor balanol.
J.Med.Chem., 53, 2010
|
|
4DPZ
 
 | | Crystal structure of human HRASLS2 | | Descriptor: | HRAS-like suppressor 2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-14 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
4DOT
 
 | | Crystal structure of human HRASLS3. | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5HI9
 
 | | Structure of the full-length TRPV2 channel by cryo-electron microscopy | | Descriptor: | Transient Receptor Potential Cation Channel Subfamily V Member 2 | | Authors: | Huynh, K.W, Cohen, M.R, Jiansen, J, Samanta, A, Lodowski, D.T, Zhou, Z.H, Moiseenkova-Bell, V.Y. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the full-length TRPV2 channel by cryo-EM.
Nat Commun, 7, 2016
|
|
2I35
 
 | | Crystal structure of rhombohedral crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I36
 
 | | Crystal structure of trigonal crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, Rhodopsin, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5KWM
 
 | |
5DJ7
 
 | |
5DKM
 
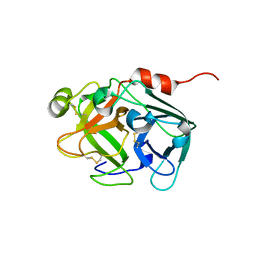 | |
5DK1
 
 | |
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4YEU
 
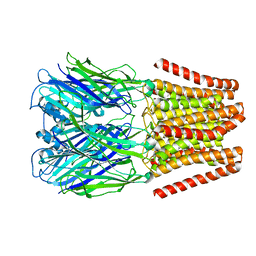 | |
6N1G
 
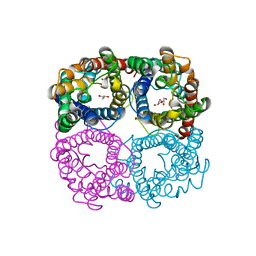 | |
3KRW
 
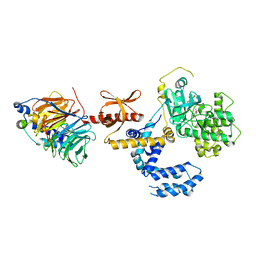 | | Human GRK2 in complex with Gbetgamma subunits and balanol (soak) | | Descriptor: | BALANOL, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tesmer, J.J.G, Tesmer, V.M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human g protein-coupled receptor kinase 2 in complex with the kinase inhibitor balanol.
J.Med.Chem., 53, 2010
|
|
3KRX
 
 | |
3MZS
 
 | | Crystal Structure of Cytochrome P450 CYP11A1 in complex with 22-hydroxy-cholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Cholesterol side-chain cleavage enzyme, ISOPROPYL ALCOHOL, ... | | Authors: | Stout, C.D, Annalora, A, Mast, N, Pikuleva, I. | | Deposit date: | 2010-05-12 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Three-step Sequential Catalysis by the Cholesterol Side Chain Cleavage Enzyme CYP11A1.
J.Biol.Chem., 286, 2011
|
|
