4C56
 
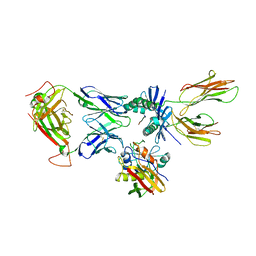 | | X-ray structure of the complex between staphylococcal enterotoxin B, T cell receptor and major histocompatibility complex class II | | Descriptor: | ENTEROTOXIN B, GLYCEROL, HEMAGGLUTININ, ... | | Authors: | Rodstrom, K.E.J, Elbing, K, Lindkvist-Petersson, K. | | Deposit date: | 2013-09-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Superantigen Staphylococcal Enterotoxin B in Complex with Tcr and Peptide-Mhc Demonstrates Absence of Tcr-Peptide Contacts.
J.Immunol., 193, 2014
|
|
6FGO
 
 | | Fc in complex with engineered calcium binding domain Z | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Venskutonyte, R, Kanje, S, Hober, S, Lindkvist-Petersson, K. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Engineering Allows for Mild Affinity-based Elution of Therapeutic Antibodies.
J. Mol. Biol., 430, 2018
|
|
4UDU
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | ENTEROTOXIN TYPE E, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
5FK9
 
 | |
8AMX
 
 | | AQP7 dimer of tetramers_D4 | | Descriptor: | Aquaporin-7 | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8AMW
 
 | | AQP7 dimer of tetramers_C1 | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Huang, P, Venskutonyte, R, Fan, X, Li, P, Yan, N, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure supports a role of AQP7 as a junction protein.
Nat Commun, 14, 2023
|
|
8C9H
 
 | | AQP7_inhibitor | | Descriptor: | Aquaporin-7, ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate | | Authors: | Huang, P, Venskutonyte, R, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2023-01-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for human aquaporin inhibition.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2W1P
 
 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | Descriptor: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-20 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
2W2E
 
 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
7ZQJ
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P3 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
7ZQI
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
6QZJ
 
 | | Crystal structure of human Aquaporin 7 at 2.2 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6QZI
 
 | | Crystal structure of human Aquaporin 7 at 1.9 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6T86
 
 | | Urocanate reductase in complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T87
 
 | | Urocanate reductase in complex with urocanate | | Descriptor: | (2E)-3-(1H-IMIDAZOL-4-YL)ACRYLIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T88
 
 | |
6T85
 
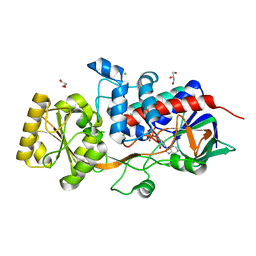 | | Urocanate reductase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
2XNA
 
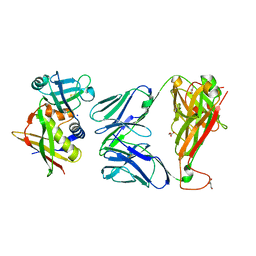 | | Crystal structure of the complex between human T cell receptor and staphylococcal enterotoxin | | Descriptor: | ENTEROTOXIN H, GLYCEROL, SODIUM ION, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
2XN9
 
 | | Crystal structure of the ternary complex between human T cell receptor, staphylococcal enterotoxin H and human major histocompatibility complex class II | | Descriptor: | ENTEROTOXIN H, GLYCEROL, HEMAGGLUTININ, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
7PGE
 
 | | copper transporter PcoB | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Copper resistance protein B, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, P, Gourdon, P.E. | | Deposit date: | 2021-08-13 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PcoB is a defense outer membrane protein that facilitates cellular uptake of copper.
Protein Sci., 31, 2022
|
|
7PRU
 
 | |
7PSD
 
 | |
7PRO
 
 | |
