2JPB
 
 | |
2JPC
 
 | |
1S7E
 
 | | Solution structure of HNF-6 | | Descriptor: | Hepatocyte nuclear factor 6 | | Authors: | Liao, X, Sheng, W. | | Deposit date: | 2004-01-29 | | Release date: | 2004-12-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA.
J.Biol.Chem., 279, 2004
|
|
1KQ8
 
 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
2HDC
 
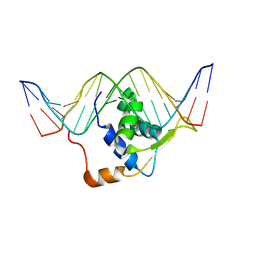 | | STRUCTURE OF TRANSCRIPTION FACTOR GENESIS/DNA COMPLEX | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*AP*AP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*TP*TP*AP*AP*GP*C)-3'), PROTEIN (TRANSCRIPTION FACTOR) | | Authors: | Jin, C, Marsden, I, Chen, X, Liao, X. | | Deposit date: | 1999-05-05 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex.
J.Mol.Biol., 289, 1999
|
|
2HFH
 
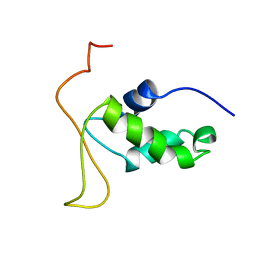 | |
1L2N
 
 | | Smt3 Solution Structure | | Descriptor: | Ubiquitin-like protein SMT3 | | Authors: | Sheng, W, Liao, X. | | Deposit date: | 2002-02-22 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions.
Protein Sci., 11, 2002
|
|
5IE8
 
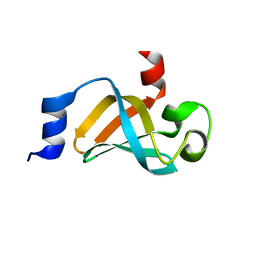 | |
7E5W
 
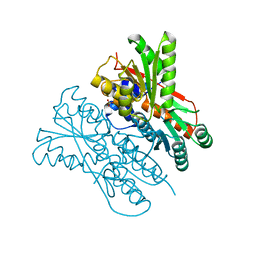 | | The structure of CcpA from Staphylococcus aureus | | Descriptor: | Catabolite control protein A, SULFATE ION | | Authors: | Yu, G, Wei, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Regulation of DNA-binding activity of the Staphylococcus aureus catabolite control protein A by copper (II)-mediated oxidation.
J.Biol.Chem., 298, 2022
|
|
1BV8
 
 | |
4NNG
 
 | |
4NNH
 
 | |
4NNI
 
 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
4NNK
 
 | |
4XSQ
 
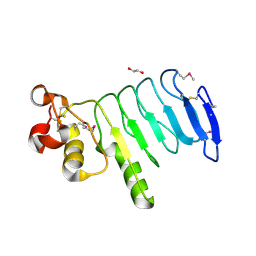 | | Structure of a variable lymphocyte receptor-like protein Bf66946 from Branchiostoma floridae | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, variable lymphocyte receptor-like protein Bf66946 | | Authors: | Cao, D.D, Cheng, W, Jiang, Y.L, Wang, W.J, Li, Q, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of a variable lymphocyte receptor-like protein from the amphioxus Branchiostoma floridae.
Sci Rep, 6, 2016
|
|
7XS8
 
 | |
7XSA
 
 | |
7XSB
 
 | |
7XSC
 
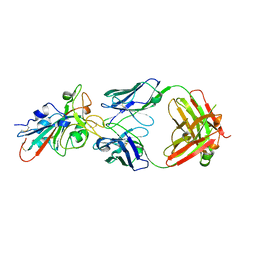 | |
7EAG
 
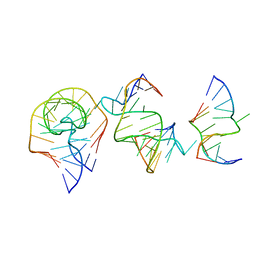 | | Crystal structure of the RAGATH-18 k-turn | | Descriptor: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
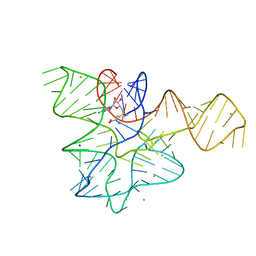 | |
5Y66
 
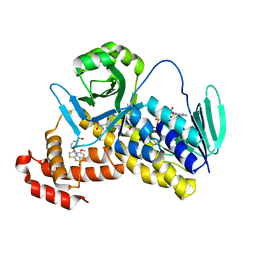 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5Y7A
 
 | |
5Y77
 
 | |
7CQ1
 
 | |
