1C9A
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-11-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
1C98
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-08-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
6N8T
 
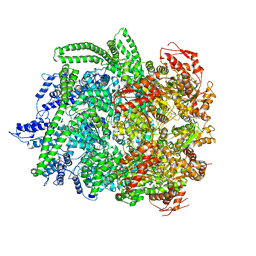 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
8JZH
 
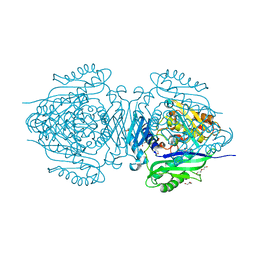 | | C. glutamicum S-adenosylmethionine synthase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, S-adenosylmethionine synthase, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZI
 
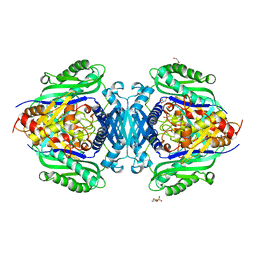 | | Mutant S-adenosylmethionine synthase from C. glutamicum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZG
 
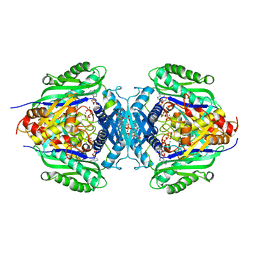 | | C. glutamicum S-adenosylmethionine synthase co-crystallized with Adenosine, triphosphate, and SAM | | Descriptor: | ADENOSINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8DEH
 
 | | Ankyrin domain of SKD3 | | Descriptor: | Caseinolytic peptidase B protein homolog | | Authors: | Lee, S, Tsai, F.T.F. | | Deposit date: | 2022-06-20 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria
Nat Commun, 14, 2023
|
|
8FDS
 
 | | Ankyrin domain of SKD3 isoform 2 | | Descriptor: | Caseinolytic peptidase B protein homolog, FORMIC ACID | | Authors: | Lee, S, Tsai, F.T.F, Chang, C. | | Deposit date: | 2022-12-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria
Nat Commun, 14, 2023
|
|
1SVP
 
 | | SINDBIS VIRUS CAPSID PROTEIN | | Descriptor: | SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Rossmann, M.G. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly.
Structure, 4, 1996
|
|
1WYK
 
 | | SINDBIS VIRUS CAPSID PROTEIN (114-264) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FORMYL GROUP, SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the potential glycoprotein binding site of sindbis virus capsid protein with dioxane and model building.
Proteins, 33, 1998
|
|
3D4V
 
 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
1P7G
 
 | | Crystal structure of superoxide dismutase from Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Superoxide dismutase | | Authors: | Lee, S, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2003-05-01 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of superoxide dismutase from Pyrobaculum aerophilum presents a challenging case in molecular replacement with multiple molecules, pseudo-symmetry and twinning.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QVR
 
 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
5IP5
 
 | |
2KM9
 
 | | Omega conotoxin-FVIA | | Descriptor: | omega_conotoxin-FVIA | | Authors: | Lee, S, Kim, J, Lee, J, Jung, H. | | Deposit date: | 2009-07-25 | | Release date: | 2010-07-28 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Structure of omega conotoxin-FVIA
To be Published
|
|
3HAK
 
 | | Human prion protein variant V129 | | Descriptor: | Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-01 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HAF
 
 | | Human prion protein variant V129 domain swapped dimer | | Descriptor: | CADMIUM ION, CHLORIDE ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-01 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HEQ
 
 | | Human prion protein variant D178N with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HES
 
 | | Human prion protein variant F198S with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HJX
 
 | | Human prion protein variant D178N with V129 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-22 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HER
 
 | | Human prion protein variant F198S with V129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HJ5
 
 | | Human prion protein variant V129 domain swapped dimer | | Descriptor: | Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-20 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HR8
 
 | | Crystal Structure of Thermotoga maritima RecA | | Descriptor: | Protein recA | | Authors: | Lee, S, Kim, T.G, Jeong, E.-Y, Ban, C, Jeon, W.-J, Min, K.I, Song, K.-M, Heo, S.-D, Ku, J.K. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of RecA Protein from Thermotoga maritima MSB8
to be published
|
|
3CRC
 
 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
3CRA
 
 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
