2QI2
 
 | | Crystal structure of the Thermoplasma acidophilum Pelota protein | | Descriptor: | Cell division protein pelota related protein | | Authors: | Lee, H.H, Kim, Y.S, Kim, K.H, Heo, I.H, Kim, S.K, Kim, O, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into dom34, a key component of no-go mRNA decay
Mol.Cell, 27, 2007
|
|
1WOR
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, DIHYDROLIPOIC ACID | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOP
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOS
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
1WOO
 
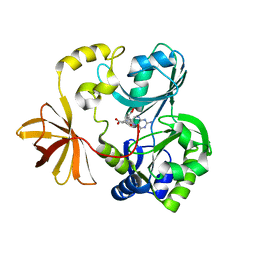 | | Crystal structure of T-protein of the Glycine Cleavage System | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Aminomethyltransferase | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
3OBY
 
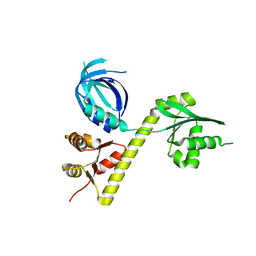 | | Crystal structure of Archaeoglobus fulgidus Pelota reveals inter-domain structural plasticity | | Descriptor: | Protein pelota homolog | | Authors: | Lee, H.H, Jang, J.Y, Yoon, H.-J, Kim, S.J, Suh, S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two archaeal Pelotas reveal inter-domain structural plasticity
Biochem.Biophys.Res.Commun., 399, 2010
|
|
3OBW
 
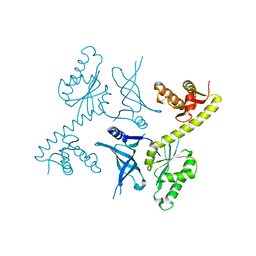 | | Crystal structure of two archaeal Pelotas reveal inter-domain structural plasticity | | Descriptor: | Protein pelota homolog | | Authors: | Lee, H.H, Jang, J.Y, Yoon, H.-J, Kim, S.J, Suh, S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two archaeal Pelotas reveal inter-domain structural plasticity
Biochem.Biophys.Res.Commun., 399, 2010
|
|
2P1B
 
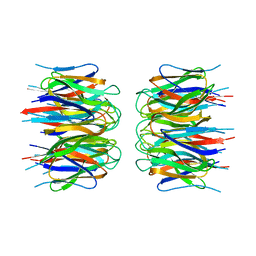 | | Crystal structure of human nucleophosmin-core | | Descriptor: | Nucleophosmin | | Authors: | Lee, H.H, Kim, H.S, Kang, J.Y, Lee, B.I, Ha, J.Y, Yoon, H.J, Lim, S.O, Jung, G, Suh, S.W. | | Deposit date: | 2007-03-03 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface
Proteins, 69, 2007
|
|
3F3M
 
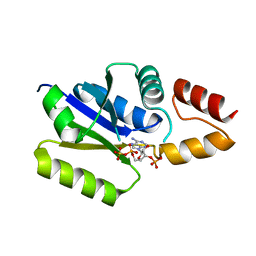 | |
2PD3
 
 | | Crystal Structure of the Helicobacter pylori Enoyl-Acyl Carrier Protein Reductase in Complex with Hydroxydiphenyl Ether Compounds, Triclosan and Diclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Lee, H.H, Moon, J.H, Suh, S.W. | | Deposit date: | 2007-03-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Helicobacter pylori enoyl-acyl carrier protein reductase in complex with hydroxydiphenyl ether compounds, triclosan and diclosan
Proteins, 69, 2007
|
|
2PD4
 
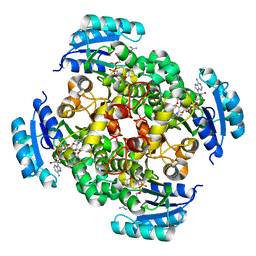 | | Crystal Structure of the Helicobacter pylori Enoyl-Acyl Carrier Protein Reductase in Complex with Hydroxydiphenyl Ether Compounds, Triclosan and Diclosan | | Descriptor: | DICLOSAN, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, H.H, Moon, J.H, Suh, S.W. | | Deposit date: | 2007-03-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Helicobacter pylori enoyl-acyl carrier protein reductase in complex with hydroxydiphenyl ether compounds, triclosan and diclosan
Proteins, 69, 2007
|
|
3E1R
 
 | | Midbody targeting of the ESCRT machinery by a non-canonical coiled-coil in CEP55 | | Descriptor: | Centrosomal protein of 55 kDa, Programmed cell death 6-interacting protein | | Authors: | Lee, H.H, Elia, N, Ghirlando, R, Lippincott-Schwartz, J, Hurley, J.H. | | Deposit date: | 2008-08-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Science, 322, 2008
|
|
2F5G
 
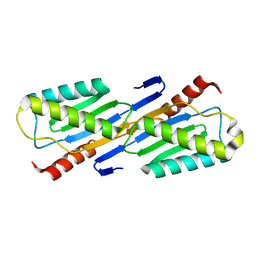 | | Crystal structure of IS200 transposase | | Descriptor: | Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2F4F
 
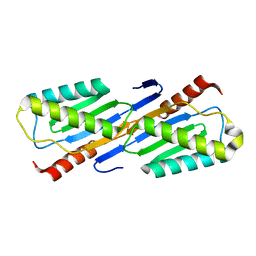 | | Crystal structure of IS200 transposase | | Descriptor: | MANGANESE (II) ION, Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
3U62
 
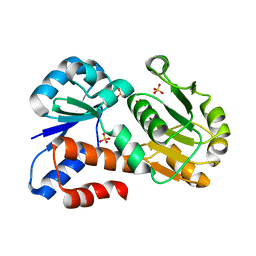 | |
8X9F
 
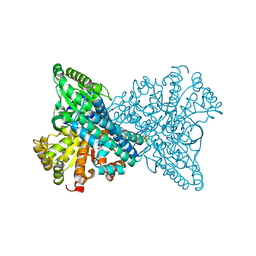 | | Crystal structure of CO dehydrogenase mutant in complex with EV | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-4-(1-ethylpyridin-1-ium-4-yl)pyridin-1-ium, Carbon monoxide dehydrogenase 2, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity
Nat Commun, 15, 2024
|
|
8X9E
 
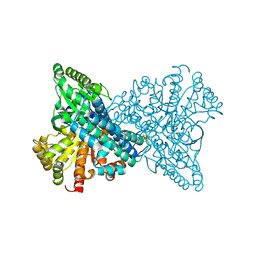 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in low PEG concentration | | Descriptor: | 1,2-ETHANEDIOL, Carbon monoxide dehydrogenase 2, FE (III) ION, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity
Nat Commun, 15, 2024
|
|
8X9G
 
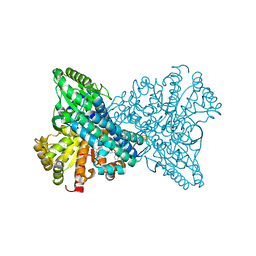 | | Crystal structure of CO dehydrogenase mutant in complex with BV | | Descriptor: | 1-(phenylmethyl)-4-[1-(phenylmethyl)pyridin-1-ium-4-yl]pyridin-1-ium, Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity
Nat Commun, 15, 2024
|
|
8X9H
 
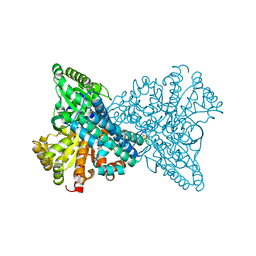 | | Crystal structure of CO dehydrogenase mutant (F41C) | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity
Nat Commun, 15, 2024
|
|
8X9D
 
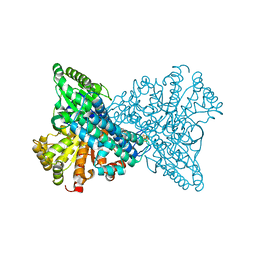 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in high PEG concentration | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity
Nat Commun, 15, 2024
|
|
5IPW
 
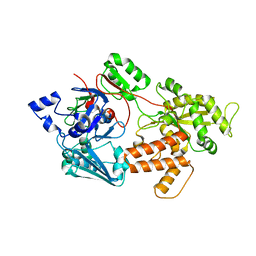 | | oligopeptide-binding protein OppA | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, putative | | Authors: | Lee, H.H, Kim, H.J, Yoon, H.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a putative oligopeptide-binding periplasmic protein from a hyperthermophile
Extremophiles, 20, 2016
|
|
5IMJ
 
 | |
8HKP
 
 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry) | | Descriptor: | Gap junction delta-2 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-11-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
2ARU
 
 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ARS
 
 | | Crystal structure of lipoate-protein ligase A From Thermoplasma acidophilum | | Descriptor: | Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
