1YG0
 
 | |
7VHV
 
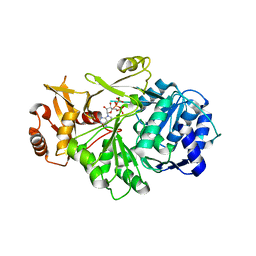 | | Crystal structure of S. aureus D-alanine alanyl carrier protein ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanyl carrier protein ligase, MAGNESIUM ION | | Authors: | Lee, B.J, Lee, I.-G, Im, H.G, Yoon, H.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of the D-alanyl carrier protein ligase DltA from Staphylococcus aureus Mu50.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4Z1A
 
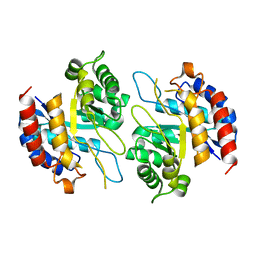 | | Structure of apo form KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1D
 
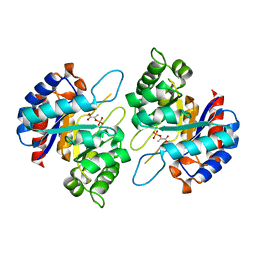 | | Structure of PEP and zinc bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE, ZINC ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1C
 
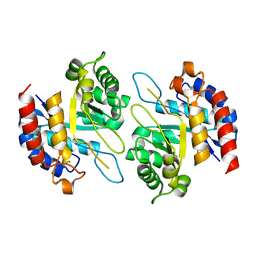 | | Structure of Cadmium bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1B
 
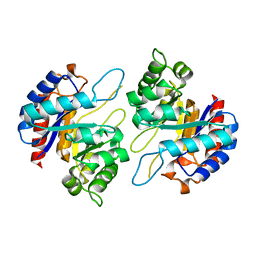 | | Structure of H204A mutant KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4N9H
 
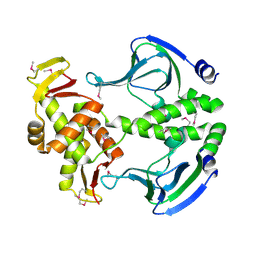 | | Crystal structure of Transcription regulation Protein CRP | | Descriptor: | Catabolite gene activator | | Authors: | Lee, B.J, Seok, S.H, Im, H, Yoon, H.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LTT
 
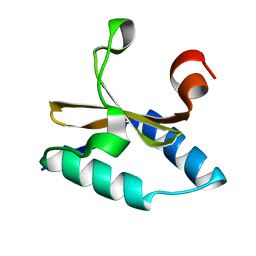 | | Crystal structure of native apo toxin from Helicobacter pylori | | Descriptor: | Uncharacterized protein, toxin | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4LSY
 
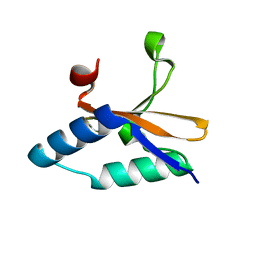 | | Crystal structure of copper-bound L66S mutant toxin from Helicobacter pylori | | Descriptor: | CITRATE ANION, COPPER (II) ION, Uncharacterized protein, ... | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4NRN
 
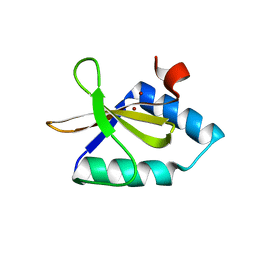 | | Crystal structure of metal-bound toxin from Helicobacter pylori | | Descriptor: | ZINC ION, metal-bound toxin | | Authors: | Lee, B.J, Im, H, Pathak, C, Jang, S.B. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of toxin HP0892 from Helicobacter pylori with two Zn(II) at 1.8 angstrom resolution
Protein Sci., 23, 2014
|
|
4Y1E
 
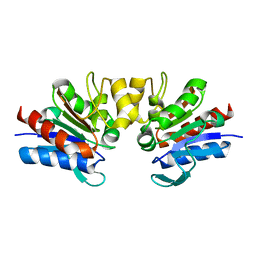 | | SAV1875-C105D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
8H2D
 
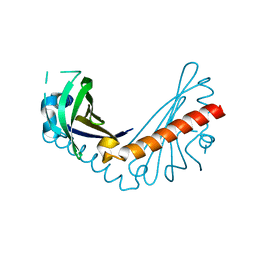 | |
8H0H
 
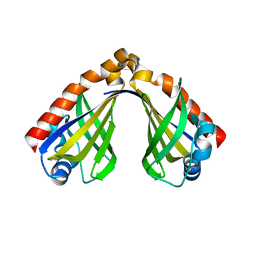 | |
3U7K
 
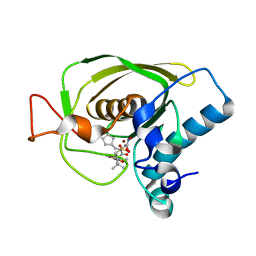 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-N-(2-(hydroxyamino)-2-oxoethyl)-2-(3-(2-methoxyphenyl)ureido)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7N
 
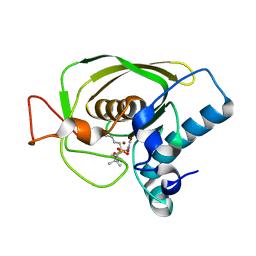 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-5-methyl-4-(3-(5-methylpyridin-2-yl)ureido)-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7L
 
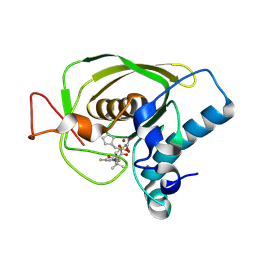 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-2-(3-(3,5-difluorophenyl)ureido)-N-(2-(hydroxyamino)-2-oxoethyl)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7M
 
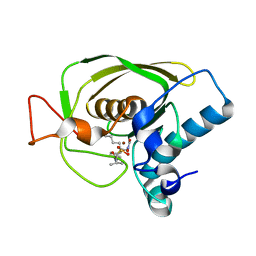 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-4-(3-(2-fluorophenyl)ureido)-5-methyl-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5HS9
 
 | |
5HS7
 
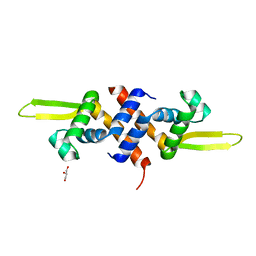 | |
5HS8
 
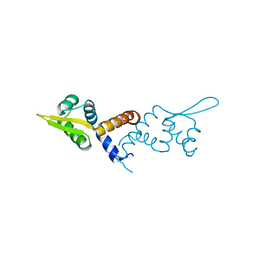 | |
3UI3
 
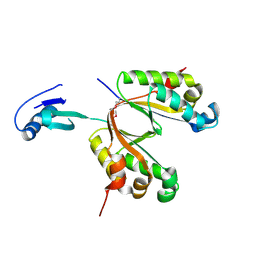 | |
4Z9E
 
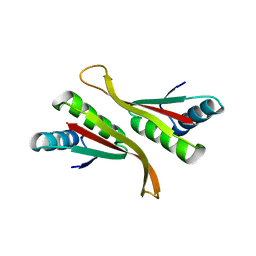 | | Alba from Thermoplasma volcanium | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Ma, C, Lee, S.J, Pathak, C, Lee, B.J. | | Deposit date: | 2015-04-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Alba from Thermoplasma volcanium belongs to alpha-NAT's: An insight into the structural aspects of Tv Alba and its acetylation by Tv Ard1.
Arch.Biochem.Biophys., 590, 2016
|
|
6IFM
 
 | | Crystal structure of DNA bound VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, DNA backward (27-MER), DNA forward (27-MER), ... | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
6IFC
 
 | | Crystal structure of VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, CALCIUM ION, tRNA(fMet)-specific endonuclease VapC | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
4PV6
 
 | | Crystal Structure Analysis of Ard1 from Thermoplasma volcanium | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-terminal acetyltransferase complex subunit [ARD1] | | Authors: | Ma, C, Lee, S.J, Lee, B.J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A.
Biochim.Biophys.Acta, 1844, 2014
|
|
