3C1U
 
 | | D192N mutant of Rhamnogalacturonan acetylesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Rhamnogalacturonan acetylesterase | | Authors: | Langkilde, A, Lo Leggio, L, Navarro Poulsen, J.C, Molgaard, A, Larsen, S. | | Deposit date: | 2008-01-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Short strong hydrogen bonds in proteins: a case study of rhamnogalacturonan acetylesterase
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
1PP4
 
 | |
7P6M
 
 | | Hydrogenated refolded hen egg-white lysozyme | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4CAG
 
 | | Bacillus licheniformis Rhamnogalacturonan Lyase PL11 | | Descriptor: | CALCIUM ION, GLYCEROL, POLYSACCHARIDE LYASE FAMILY 11 PROTEIN | | Authors: | Otten, H, Rodrigues da Silva, I.I.C, Jers, C, Nyffenegger, C, Larsen, D.M, Mikkelsen, J.D, Larsen, S. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Design of Thermostable Rhamnogalacturonan Lyase Mutants from Bacillus Licheniformis by Combination of Targeted Single Point Mutations.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
1RMG
 
 | | RHAMNOGALACTURONASE A FROM ASPERGILLUS ACULEATUS | | Descriptor: | RHAMNOGALACTURONASE A, alpha-D-glucopyranose, alpha-D-mannopyranose, ... | | Authors: | Petersen, T.N, Kauppinen, S, Larsen, S. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix.
Structure, 5, 1997
|
|
1JQV
 
 | | The K213E mutant of Lactococcus lactis Dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRB
 
 | | The P56A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JUB
 
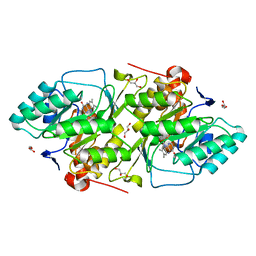 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JQX
 
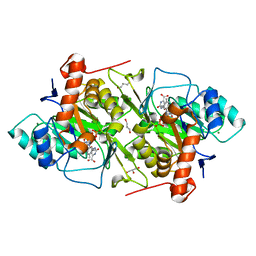 | | The R57A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRC
 
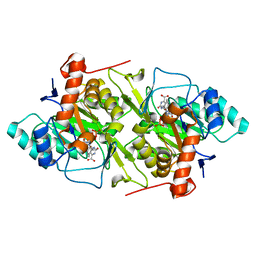 | | The N67A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JUE
 
 | | 1.8 A resolution structure of native lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1DEO
 
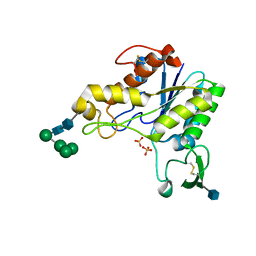 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, SULFATE ION, ... | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-15 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
1DEX
 
 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.9 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-16 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
1NKG
 
 | | Rhamnogalacturonan lyase from Aspergillus aculeatus | | Descriptor: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION | | Authors: | McDonough, M.A, Kadirvelraj, R, Harris, P, Poulsen, J.C, Larsen, S. | | Deposit date: | 2003-01-03 | | Release date: | 2004-05-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rhamnogalacturonan lyase reveals a unique three-domain modular structure for polysaccharide lyase family 4.
Febs Lett., 565, 2004
|
|
1DKR
 
 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DKU
 
 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | METHYL PHOSPHONIC ACID ADENOSINE ESTER, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN (PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE) | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
3NJX
 
 | | Rhamnogalacturonan Lyase from Aspergillus aculeatus mutant H210A | | Descriptor: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION | | Authors: | Jensen, M.H, Otten, H, Christensen, U, Borchert, T.V, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Studies Elucidate the Mechanism of Rhamnogalacturonan Lyase from Aspergillus aculeatus.
J.Mol.Biol., 404, 2010
|
|
3NJV
 
 | | Rhamnogalacturonan lyase from Aspergillus aculeatus K150A substrate complex | | Descriptor: | CALCIUM ION, Rhamnogalacturonase B, SULFATE ION, ... | | Authors: | Jensen, M.H, Otten, H, Christensen, U, Borchert, T.V, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Studies Elucidate the Mechanism of Rhamnogalacturonan Lyase from Aspergillus aculeatus.
J.Mol.Biol., 404, 2010
|
|
1OCX
 
 | | E. coli maltose-O-acetyltransferase | | Descriptor: | MALTOSE O-ACETYLTRANSFERASE, TRIMETHYL LEAD ION | | Authors: | Lo Leggio, L, Dal Degan, F, Poulsen, P, Larsen, S. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Specificity of Escherichia Coli Maltose Acetyltransferase Give New Insight Into the Laca Family of Acyltransferases.
Biochemistry, 42, 2003
|
|
1FHL
 
 | |
1FOB
 
 | |
2G3N
 
 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA in complex with beta-octyl-glucopyranoside | | Descriptor: | Alpha-glucosidase, octyl beta-D-glucopyranoside | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
