2LBO
 
 | |
2OCI
 
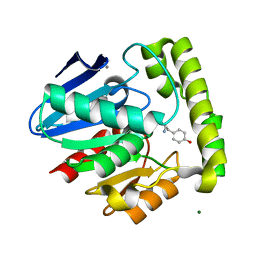 | | Crystal structure of valacyclovir hydrolase complexed with a product analogue | | Descriptor: | L-TYROSINAMIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCK
 
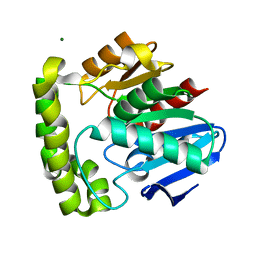 | | Crystal structure of valacyclovir hydrolase D123N mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCL
 
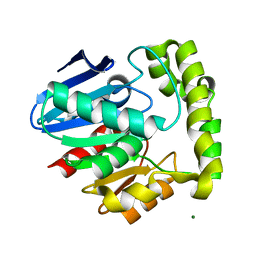 | | Crystal structure of valacyclovir hydrolase S122A mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Valacyclovir hydrolase | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
2OCG
 
 | | Crystal structure of human valacyclovir hydrolase | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lai, L, Xu, Z, Amidon, G.L. | | Deposit date: | 2006-12-20 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of prodrug activation by human valacyclovirase, an alpha-amino acid ester hydrolase.
J.Biol.Chem., 283, 2008
|
|
4A5V
 
 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
2KI0
 
 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3TKK
 
 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
7C0N
 
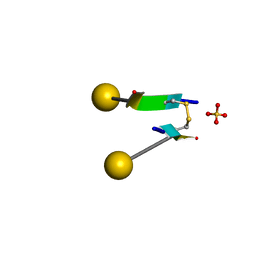 | | Crystal structure of a self-assembling galactosylated peptide homodimer | | Descriptor: | SULFATE ION, Self-assembling galactosylated tyrosine-rich peptide, beta-D-galactopyranose | | Authors: | He, C, Wu, S, Chi, C, Zhang, W, Ma, M, Lai, L, Dong, S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Glycopeptide Self-Assembly Modulated by Glycan Stereochemistry through Glycan-Aromatic Interactions.
J.Am.Chem.Soc., 142, 2020
|
|
1EKE
 
 | | CRYSTAL STRUCTURE OF CLASS II RIBONUCLEASE H (RNASE HII) WITH MES LIGAND | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, RIBONUCLEASE HII | | Authors: | Lai, L.H, Yokota, H, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-03-07 | | Release date: | 2000-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of archaeal RNase HII: a homologue of human major RNase H
Structure, 8, 2000
|
|
2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
7VLR
 
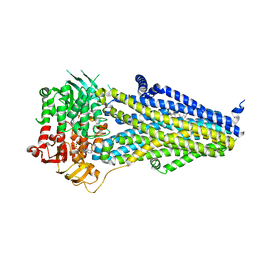 | | Structure of SUR2B in complex with Mg-ATP/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLT
 
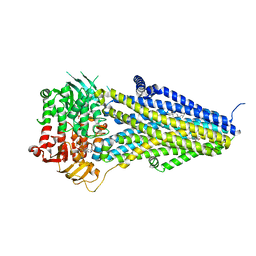 | | Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim | | Descriptor: | (3S,4R)-2,2-dimethyl-3-oxidanyl-4-(2-oxidanylidenepyrrolidin-1-yl)-3,4-dihydrochromene-6-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLS
 
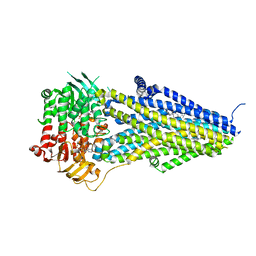 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLU
 
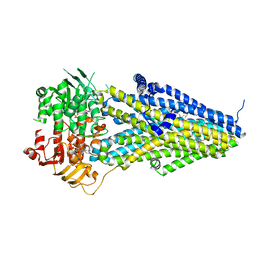 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
2LL4
 
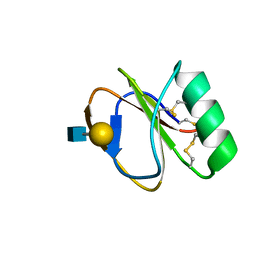 | |
2LL3
 
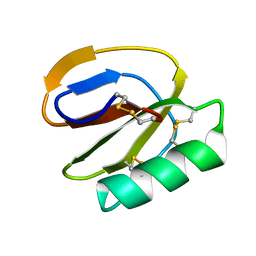 | |
4FZP
 
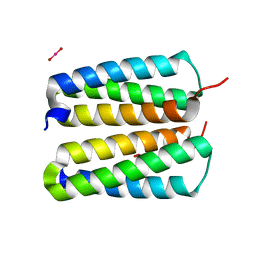 | |
4FZO
 
 | |
4TUR
 
 | |
4TUS
 
 | |
4TUQ
 
 | | Human DNA polymerase beta inserting dCMPNPP opposite GG template (GG0b). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*CP*CP*AP*CP*GP*GP*CP*CP*CP*AP*TP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*AP*TP*GP*GP*GP*C)-3'), ... | | Authors: | Koag, M.C, Lee, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Structural Basis for the Inefficient Nucleotide Incorporation Opposite Cisplatin-DNA Lesion by Human DNA Polymerase beta.
J.Biol.Chem., 289, 2014
|
|
4TUP
 
 | |
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
7U0P
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
