4V7G
 
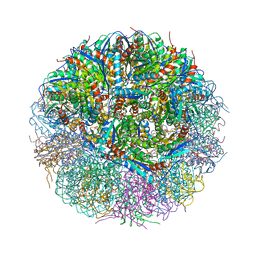 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3RJ5
 
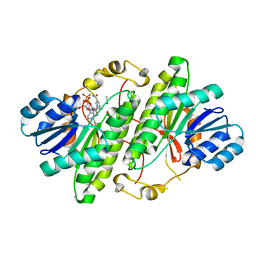 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
3RJ9
 
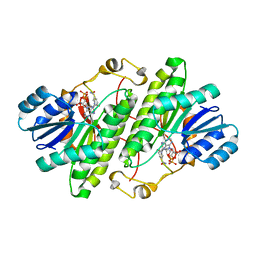 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
1MG5
 
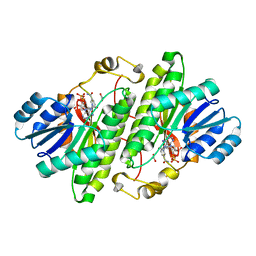 | | Crystal structure of Drosophila melanogaster alcohol dehydrogenase complexed with NADH and acetate at 1.6 A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, alcohol dehydrogenase | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 2002-08-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drosophila alcohol dehydrogenase: acetate-enzyme interactions and novel insights into the effects of electrostatics on catalysis
J.Mol.Biol., 345, 2005
|
|
3HI8
 
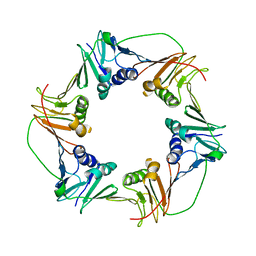 | | Crystal structure of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii | | Descriptor: | Proliferating cell nuclear antigen PcnA | | Authors: | Morgunova, E, Gray, F.C, MacNeill, S.A, Ladenstein, R. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural insights into the adaptation of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii to a high-salt environment.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1GP1
 
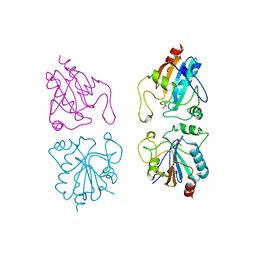 | |
1SBY
 
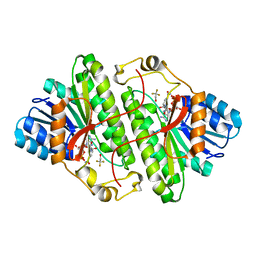 | | Alcohol dehydrogenase from Drosophila lebanonensis complexed with NAD+ and 2,2,2-trifluoroethanol at 1.1 A resolution | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL | | Authors: | Benach, J, Meijers, R, Atrian, S, Gonzalez-Duarte, R, Lamzin, V.S, Ladenstein, R. | | Deposit date: | 2004-02-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1-A crystal structure of D. lebanonensis ADH complexed with NAD+ and 2,2,2-trifluoroethanol
To be Published
|
|
2VLD
 
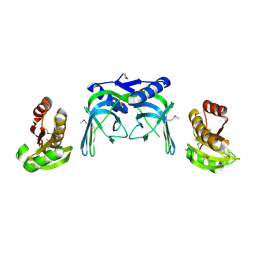 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
2IV0
 
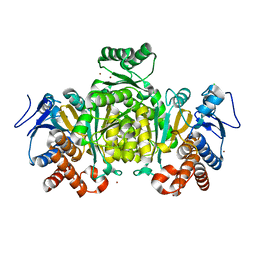 | | Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers | | Descriptor: | CHLORIDE ION, ISOCITRATE DEHYDROGENASE, ZINC ION | | Authors: | Stokke, R, Karlstrom, M, Yang, N, Leiros, I, Ladenstein, R, Birkeland, N.K, Steen, I.H. | | Deposit date: | 2006-06-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thermal Stability of Isocitrate Dehydrogenase from Archaeoglobus Fulgidus Studied by Crystal Structure Analysis and Engineering of Chimers
Extremophiles, 11, 2007
|
|
1SSO
 
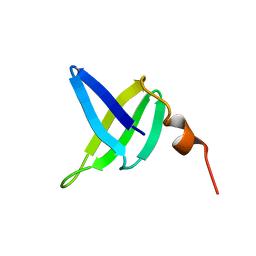 | | SOLUTION STRUCTURE AND DNA-BINDING PROPERTIES OF A THERMOSTABLE PROTEIN FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | SSO7D | | Authors: | Baumann, H, Knapp, S, Lundback, T, Ladenstein, R, Hard, T. | | Deposit date: | 1995-03-31 | | Release date: | 1995-05-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus.
Nat.Struct.Biol., 1, 1994
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
1TYO
 
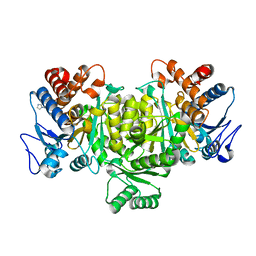 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix in complex with etheno-NADP | | Descriptor: | ETHENO-NADP, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N, Ladenstein, R. | | Deposit date: | 2004-07-08 | | Release date: | 2005-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
1JM1
 
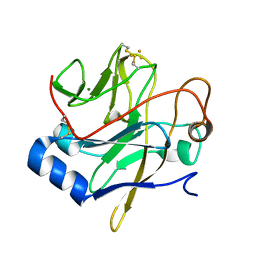 | | Crystal structure of the soluble domain of the Rieske protein II (soxF) from Sulfolobus acidocaldarius | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein soxF | | Authors: | Boenisch, H, Schmidt, C.L, Schaefer, G, Ladenstein, R. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | The structure of the soluble domain of an archaeal Rieske iron-sulfur protein at 1.1 A resolution.
J.Mol.Biol., 319, 2002
|
|
1XGV
 
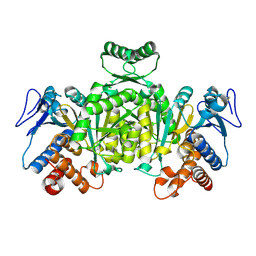 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | Isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
1B16
 
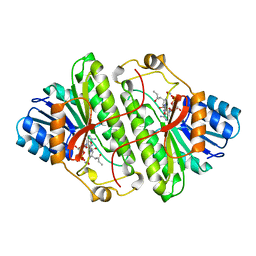 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS TERNARY COMPLEX WITH NAD-3-PENTANONE | | Descriptor: | NICOTINAMIDE ADENINE DINUCLEOTIDE 3-PENTANONE ADDUCT, PROTEIN (ALCOHOL DEHYDROGENASE) | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1B15
 
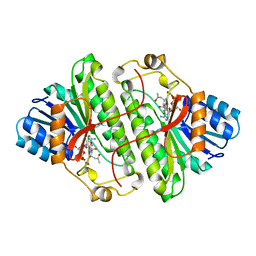 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS TERNARY COMPLEX WITH NAD-ACETONE | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE ADENINE DINUCLEOTIDE ACETONE ADDUCT | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1B2L
 
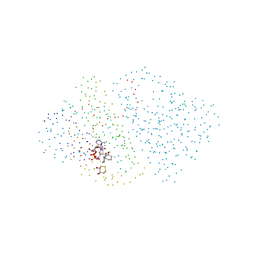 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS: TERNARY COMPLEX WITH NAD-CYCLOHEXANONE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ALCOHOL DEHYDROGENASE, CALCIUM ION, ... | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1GSS
 
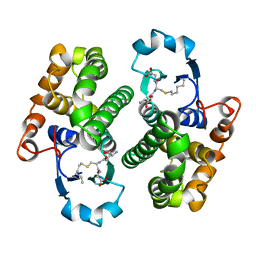 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
2P4W
 
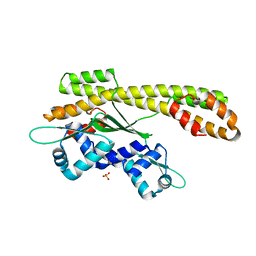 | | Crystal structure of heat shock regulator from Pyrococcus furiosus | | Descriptor: | SULFATE ION, Transcriptional regulatory protein arsR family | | Authors: | Liu, W, Vierke, G, Panjikar, S, Thomm, M, Ladenstein, R. | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Archaeal Heat Shock Regulator from Pyrococcus furiosus: A Molecular Chimera Representing Eukaryal and Bacterial Features.
J.Mol.Biol., 369, 2007
|
|
2PYA
 
 | |
1AOZ
 
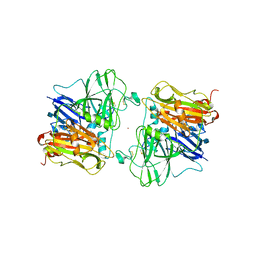 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
2JFB
 
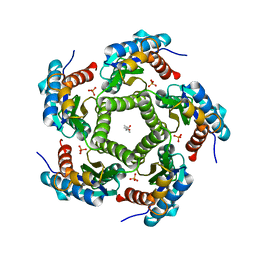 | | 3D Structure of Lumazine Synthase from Candida albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION | | Authors: | Morgunova, E, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lumazine Synthase from Candida Albicans as an Anti- Fungal Target Enzyme: Structural and Biochemical Basis for Drug Design.
J.Biol.Chem., 282, 2007
|
|
1A4U
 
 | |
2TMG
 
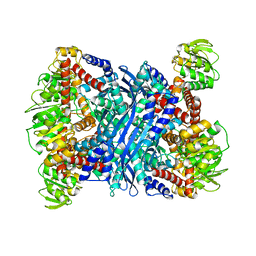 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
2VL6
 
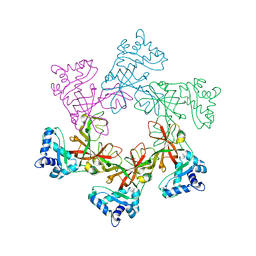 | | STRUCTURAL ANALYSIS OF THE SULFOLOBUS SOLFATARICUS MCM PROTEIN N- TERMINAL DOMAIN | | Descriptor: | MINICHROMOSOME MAINTENANCE PROTEIN MCM, ZINC ION | | Authors: | Liu, W, Pucci, B, Rossi, M, Pisani, F.M, Ladenstein, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Sulfolobus Solfataricus Mcm Protein N-Terminal Domain.
Nucleic Acids Res., 36, 2008
|
|
