1G83
 
 | | CRYSTAL STRUCTURE OF FYN SH3-SH2 | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE FYN | | Authors: | Arold, S.T, Ulmer, T.S, Mulhern, T.D, Werner, J.M, Ladbury, J.E, Campbell, I.D, Noble, M.E.M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The role of the Src homology 3-Src homology 2 interface in the regulation of Src kinases.
J.Biol.Chem., 276, 2001
|
|
6V6Q
 
 | | Crystal Structure of Monophosphorylated FGF Receptor 2 isoform IIIb with PTR657 | | Descriptor: | Fibroblast growth factor receptor 2, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lin, C.-C, Wieteska, L, Poncet-Montange, G, Suen, K.M, Arold, S.T, Ahmed, Z, Ladbury, J.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The combined action of the intracellular regions regulates FGFR2 kinase activity
Commun Biol, 6, 2023
|
|
3NR7
 
 | |
2BUG
 
 | | Solution structure of the TPR domain from Protein phosphatase 5 in complex with Hsp90 derived peptide | | Descriptor: | HSP90, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Cliff, M.J, Harris, R, Barford, D, Ladbury, J.E, Williams, M.A. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-16 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Diversity in the Tpr Domain-Mediated Interaction of Protein Phosphatase 5 with Hsp90.
Structure, 14, 2006
|
|
2O2O
 
 | |
1XF7
 
 | | High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 | | Descriptor: | Wilms' Tumor Protein, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Dong, J, Huang, K, Carey, P, Weiss, M.A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Why zinc fingers prefer zinc: ligand-field symmetry and the hidden thermodynamics of metal ion selectivity
Biochemistry, 43, 2004
|
|
1XRZ
 
 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | Descriptor: | ZINC ION, Zinc finger Y-chromosomal protein | | Authors: | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | Deposit date: | 2004-10-17 | | Release date: | 2004-11-30 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
2JYQ
 
 | | NMR structure of the apo v-Src SH2 domain | | Descriptor: | Tyrosine-protein kinase transforming protein Src | | Authors: | Taylor, J.D, Ababou, A, Williams, M.A, Ladbury, J.E. | | Deposit date: | 2007-12-17 | | Release date: | 2008-06-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: Implications for drug design
Proteins, 73, 2008
|
|
2M2D
 
 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
1KLS
 
 | | NMR Structure of the ZFY-6T[Y10L] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1KLR
 
 | | NMR Structure of the ZFY-6T[Y10F] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1B55
 
 | | PH DOMAIN FROM BRUTON'S TYROSINE KINASE IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, TYROSINE-PROTEIN KINASE BTK, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A.M, Potter, B.V.L, O'Brien, R, Ladbury, J.E, Saraste, M. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
4TT4
 
 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT2
 
 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU6
 
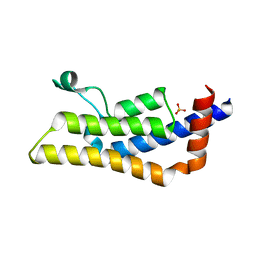 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU4
 
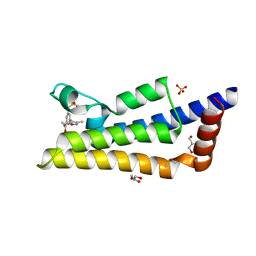 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT6
 
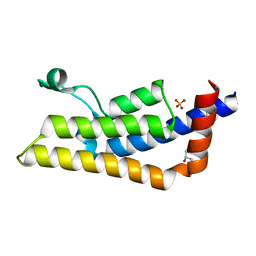 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
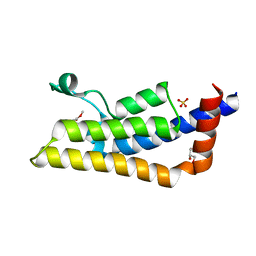 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
1QKA
 
 | |
1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | Descriptor: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
2DRU
 
 | | Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, chimera of CD48 antigen and T-cell surface antigen CD2 | | Authors: | Evans, E.J, Ikemizu, S, Davis, S.J. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Binding Properties of the CD2 and CD244 (2B4)-binding Protein, CD48
J.Biol.Chem., 281, 2006
|
|
