8S9Q
 
 | |
4TSX
 
 | |
7KE0
 
 | |
1HH1
 
 | | THE STRUCTURE OF HJC, A HOLLIDAY JUNCTION RESOLVING ENZYME FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | HOLLIDAY JUNCTION RESOLVING ENZYME HJC | | Authors: | Bond, C.S, Kvaratskhelia, M, Richard, D, White, M.F, Hunter, W.N. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hjc, a Holliday Junction Resolvase, from Sulfolobus Solfataricus
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8D3S
 
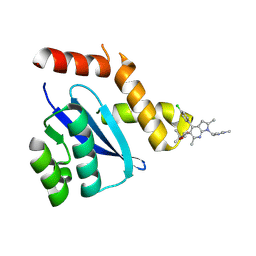 | |
6NUJ
 
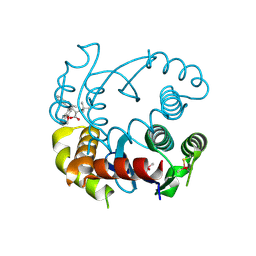 | |
4DMN
 
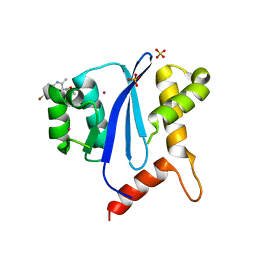 | | HIV-1 Integrase Catalytical Core Domain | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](methoxy)ethanoic acid, ARSENIC, HIV-1 Integrase, ... | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors.
J.Biol.Chem., 287, 2012
|
|
6PU1
 
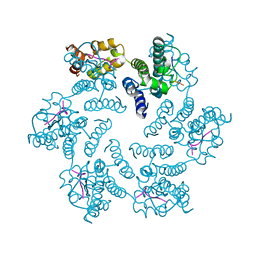 | |
6EB1
 
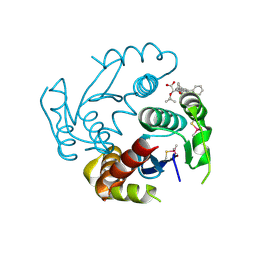 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6EB2
 
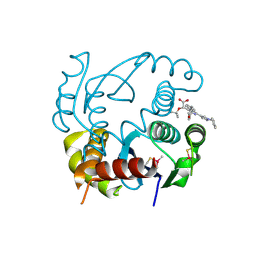 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid | | Descriptor: | (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6VKV
 
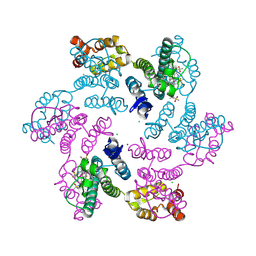 | |
6VWS
 
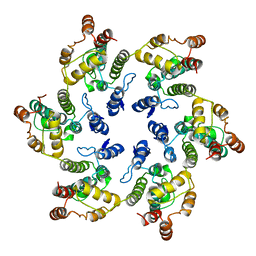 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
2V0X
 
 | | The dimerization domain of LAP2alpha | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | Authors: | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | Deposit date: | 2007-05-20 | | Release date: | 2007-06-26 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
5JL4
 
 | |
8GDV
 
 | |
7SNN
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide
To Be Published
|
|
7SNQ
 
 | |
7SNL
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide
To Be Published
|
|
4O0J
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2013-12-13 | | Release date: | 2014-07-02 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase.
Plos Pathog., 10, 2014
|
|
4O5B
 
 | |
4O55
 
 | |
4GW6
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](tert-butoxy)ethanoic acid, ARSENIC, Gag-Pol polyprotein | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The A128T Resistance Mutation Reveals Aberrant Protein Multimerization as the Primary Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors.
J.Biol.Chem., 288, 2013
|
|
4GVM
 
 | |
4ID1
 
 | |
4JLH
 
 | |
