8I7E
 
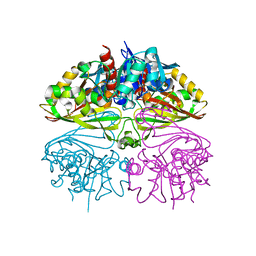 | | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Kumar, N, Dilawari, R, Chaubey, G.K, Modanwal, R, Talukdar, S, Dhiman, A, Chaudhary, S, Patidar, A, Kumar, A, Raje, C.I, Raje, M, Kumaran, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A
To Be Published
|
|
7YOG
 
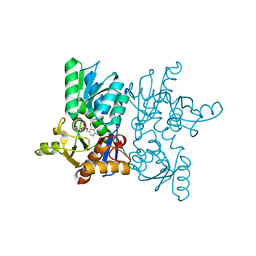 | |
7YOF
 
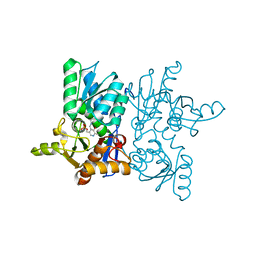 | |
7YOH
 
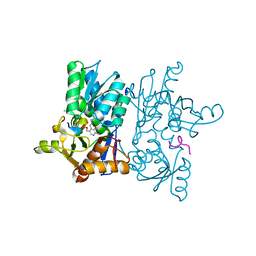 | |
7YOI
 
 | |
7YOD
 
 | |
7YOE
 
 | |
8GSK
 
 | |
6UE7
 
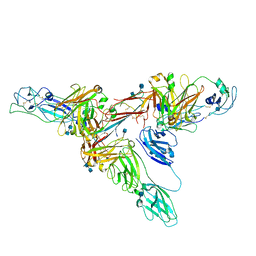 | | Structure of dimeric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UEA
 
 | | Structure of pentameric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UE8
 
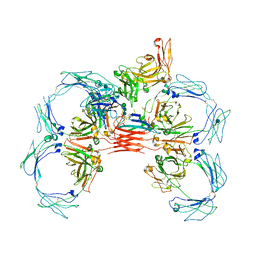 | | Structure of tetrameric sIgA complex (Class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UE9
 
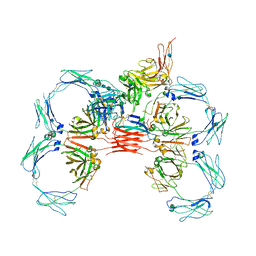 | | Structure of tetrameric sIgA complex (Class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
8WI9
 
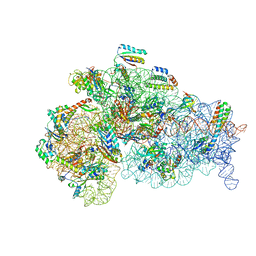 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, bS1 and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WIF
 
 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WHX
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WI7
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, bS1 and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WIB
 
 | | Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, E- tRNA and RafH. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WI8
 
 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, bS1 and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WHY
 
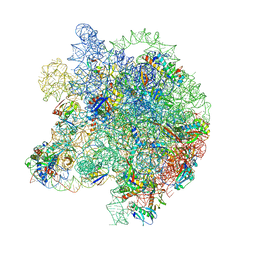 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WID
 
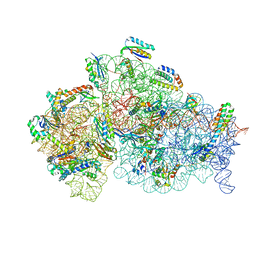 | | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, E- tRNA and RafH. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
8WIC
 
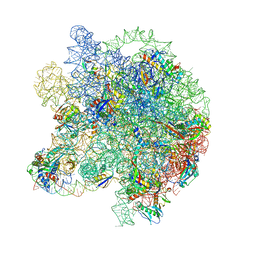 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, E- tRNA and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
7WU8
 
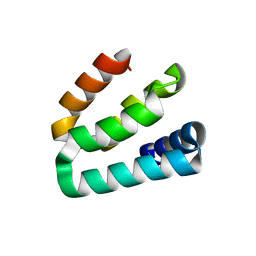 | |
6CBC
 
 | |
6K2F
 
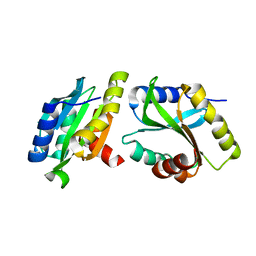 | | Structure of Eh TWINFILIN | | Descriptor: | Actin binding protein family protein | | Authors: | Kumar, N, Gourinath, S, Rath, P.P. | | Deposit date: | 2019-05-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of N-terminal Twinfilin domain from Entamoeba histolytica
To Be Published
|
|
6LCN
 
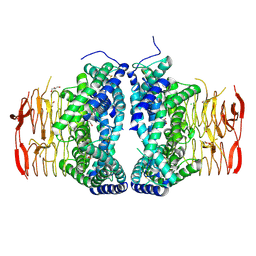 | | Crystal structure of Serine Acetyltransferase from Planctomyces limnophilus at 2.15A | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kumar, N, Singh, R.P, Singh, A.K, Kumaran, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Understanding Mechanics of competitive-allostery Using Engineered Cysteine Synthase Assembly
To Be Published
|
|
