2WX0
 
 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P21 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WX1
 
 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked tri-ubiquitin, P212121 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WWZ
 
 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P212121 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
3ZJG
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 60 min H2O2 soak. | | Descriptor: | CHLORIDE ION, TUMOR NECROSIS FACTOR ALPHA-INDUCED PROTEIN 3 | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJD
 
 | | A20 OTU domain in reduced, active state at 1.87 A resolution | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJF
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 270 min H2O2 soak. | | Descriptor: | A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJE
 
 | | A20 OTU domain in reversibly oxidised (SOH) state | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
8BZR
 
 | | UFC1-UFM1 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-fold modifier 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8C0D
 
 | | UFL1/DDRGK1 bound to UFC1 | | Descriptor: | DDRGK domain-containing protein 1, E3 UFM1-protein ligase 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
6FGE
 
 | | Crystal structure of human ZUFSP/ZUP1 in complex with ubiquitin | | Descriptor: | ACETATE ION, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kwasna, D, Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability.
Mol. Cell, 70, 2018
|
|
6TXB
 
 | |
6RUU
 
 | | Pseudokinase domain of human IRAK3 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 3, MERCURY (II) ION, ... | | Authors: | Lange, S.M, Kulathu, Y, Cohen, P. | | Deposit date: | 2019-05-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dimeric Structure of the Pseudokinase IRAK3 Suggests an Allosteric Mechanism for Negative Regulation.
Structure, 29, 2021
|
|
6TUV
 
 | |
8A67
 
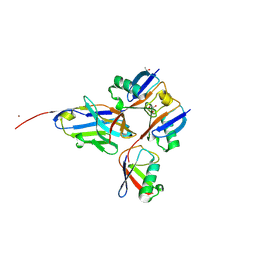 | |
4S1Z
 
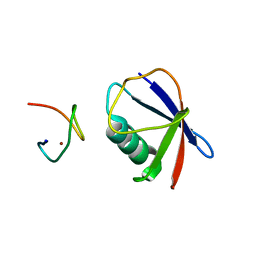 | | Crystal structure of TRABID NZF1 in complex with K29 linked di-Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin thioesterase ZRANB1, ZINC ION | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
4S22
 
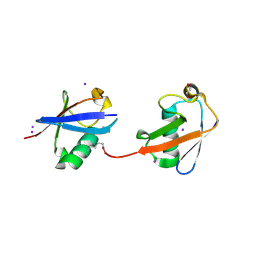 | | Crystal structure of K29 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
4Y1H
 
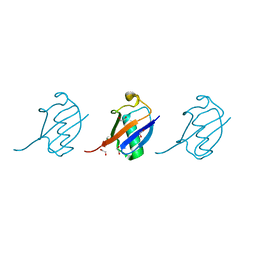 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4XYZ
 
 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
7NPI
 
 | | Crystal structure of Mindy2 (C266A) in complex with Lys48-linked penta-ubiquitin (K48-Ub5) | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, SODIUM ION, ... | | Authors: | Lange, S.M, Armstrong, L.A, Kulathu, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
7NBB
 
 | |
7NPO
 
 | | Branched K48-K63-Ub3 | | Descriptor: | GLYCEROL, Polyubiquitin-B | | Authors: | Lange, S.M, Kwasna, D, Kulathu, Y. | | Deposit date: | 2021-02-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Branched Lys48-Lys63-linked triubiquitin
To be published
|
|
3ZNV
 
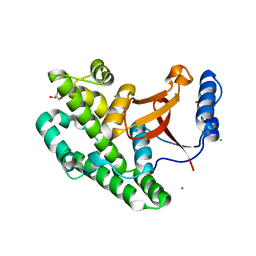 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
6Y6R
 
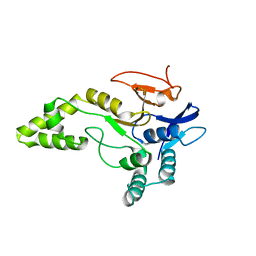 | | Crystal structure of MINDY1 T335D mutant | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6YJG
 
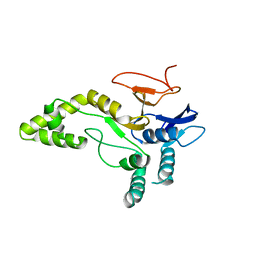 | | Crystal structure of MINDY1 mutant-Y114F | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6Z49
 
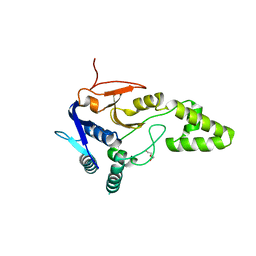 | | Crystal structure of deubiquitinase Mindy2 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-05-23 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
