4WH2
 
 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
2DRR
 
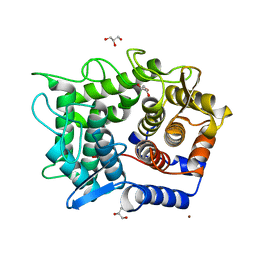 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRS
 
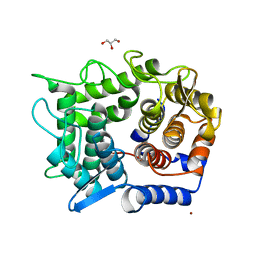 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263S mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRO
 
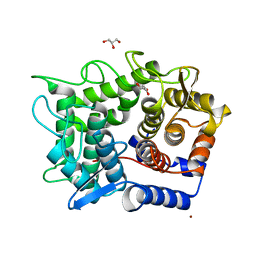 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263C mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRQ
 
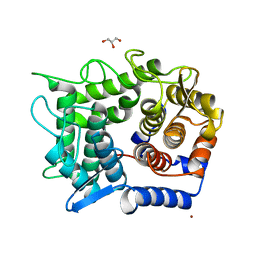 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263G mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
1WU5
 
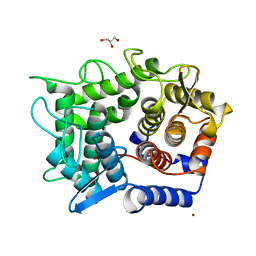 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase complexed with xylose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU4
 
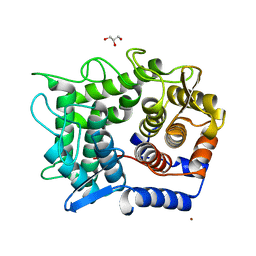 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | Descriptor: | GLYCEROL, NICKEL (II) ION, xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU6
 
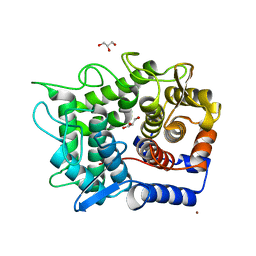 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase E70A mutant complexed with xylobiose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
2DEP
 
 | |
1V7V
 
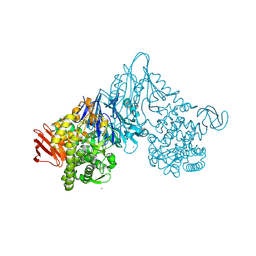 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | Descriptor: | CALCIUM ION, chitobiose phosphorylase | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7W
 
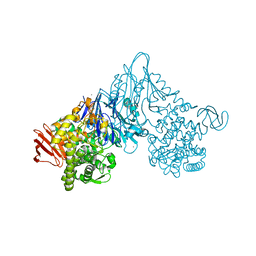 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7X
 
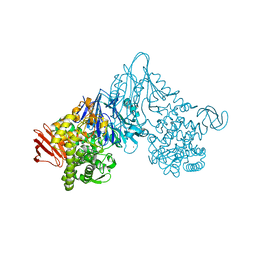 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
4UNI
 
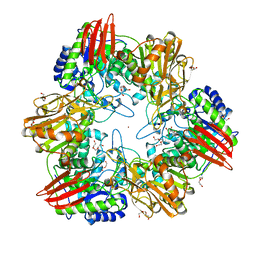 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOQ
 
 | | Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOZ
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
3QFZ
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QG0
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Cellobiose Phosphorylase, PHOSPHATE ION, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
3QFY
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Cellobiose Phosphorylase, ... | | Authors: | Fushinobu, S, Hidaka, M, Hayashi, A.M, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions between glycoside hydrolase family 94 cellobiose phosphorylase and glucosidase inhibitors
J.Appl.Glyosci., 58, 2011
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
6K0H
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-GlcNAc | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
6K0I
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-Glc | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
6K0G
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2019-05-06 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
