3BBZ
 
 | | Structure of the nucleocapsid-binding domain from the mumps virus phosphoprotein | | Descriptor: | BROMIDE ION, FORMIC ACID, P protein | | Authors: | Kingston, R.L, Gay, L.S, Baase, W.S, Matthews, B.W. | | Deposit date: | 2007-11-11 | | Release date: | 2008-05-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization
J.Mol.Biol., 379, 2008
|
|
1T6O
 
 | | Nucleocapsid-binding domain of the measles virus P protein (amino acids 457-507) in complex with amino acids 486-505 of the measles virus N protein | | Descriptor: | linker, phosphoprotein | | Authors: | Kingston, R.L, Hamel, D.J, Gay, L.S, Dahlquist, F.W, Matthews, B.W. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the attachment of a paramyxoviral polymerase to its template.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1EOQ
 
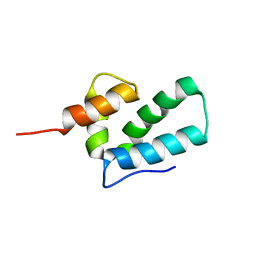 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1EM9
 
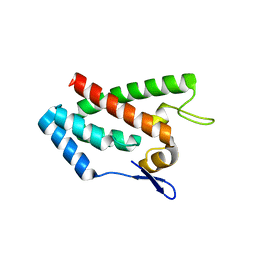 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: N-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27, MAGNESIUM ION | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1OFG
 
 | | GLUCOSE-FRUCTOSE OXIDOREDUCTASE | | Descriptor: | GLUCOSE-FRUCTOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kingston, R.L, Scopes, R.K, Baker, E.N. | | Deposit date: | 1996-10-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP.
Structure, 4, 1996
|
|
5KZA
 
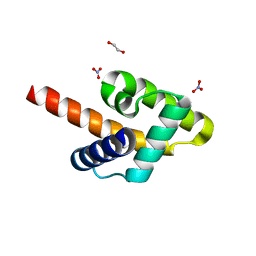 | | Crystal structure of the Rous sarcoma virus matrix protein (aa 2-102). Space group I41 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, virus matrix protein | | Authors: | Kingston, R.L, Chan, J, Vogt, V.M. | | Deposit date: | 2016-07-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties.
Biophys. J., 113, 2017
|
|
5KZ9
 
 | |
5KZB
 
 | |
3G0V
 
 | |
3G21
 
 | |
3G28
 
 | |
3G1G
 
 | |
3G29
 
 | |
3G1I
 
 | |
3G26
 
 | |
7KD4
 
 | |
7KD5
 
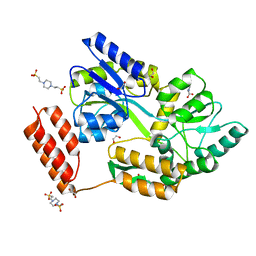 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein (residues 329 -388), fused to MBP. Space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein and Phosphoprotein fusion protein, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Webby, M.N, Kingston, R.L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions.
Viruses, 13, 2021
|
|
3TIR
 
 | |
1AZC
 
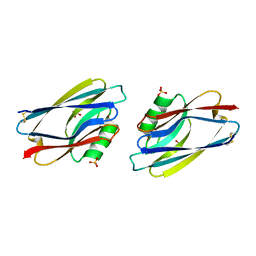 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Baker, E.N, Shepard, W.E.B, Kingston, R.L. | | Deposit date: | 1992-12-16 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2X8Q
 
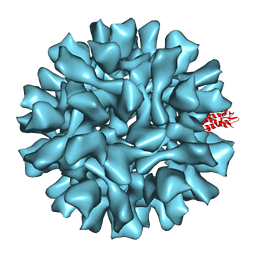 | |
1JMY
 
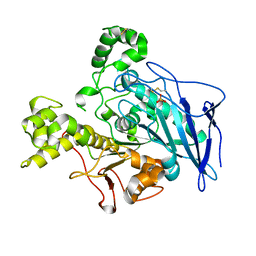 | | Truncated Recombinant Human Bile Salt Stimulated Lipase | | Descriptor: | BILE-SALT-ACTIVATED LIPASE, SULFATE ION | | Authors: | Moore, S.A, Kingston, R.L, Loomes, K.M, Hernell, O, Blackberg, L, Baker, H.M, Baker, E.N. | | Deposit date: | 2001-07-20 | | Release date: | 2001-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of truncated recombinant human bile salt-stimulated lipase reveals bile salt-independent conformational flexibility at the active-site loop and provides insights into heparin binding.
J.Mol.Biol., 312, 2001
|
|
4KYD
 
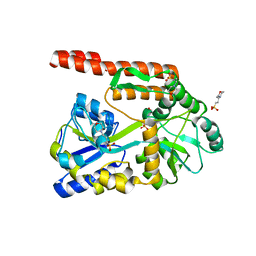 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Maltose-binding periplasmic protein, Phosphoprotein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
4KYC
 
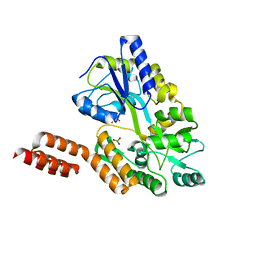 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein, fused to MBP. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Maltose-binding periplasmic protein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
4KYE
 
 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | Maltose-binding periplasmic protein, Phosphoprotein, chimeric construct, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
4P4T
 
 | | GDP-bound stalkless-MxA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Interferon-induced GTP-binding protein Mx1, SULFATE ION | | Authors: | Rennie, M.L, McKelvie, S.A, Bulloch, E.M, Kingston, R.L. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transient Dimerization of Human MxA Promotes GTP Hydrolysis, Resulting in a Mechanical Power Stroke.
Structure, 22, 2014
|
|
