4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
4X98
 
 | | Immunoglobulin Fc heterodimer variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
4X99
 
 | | Immunoglobulin Fc heterodimers variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
1DBO
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
2QI2
 
 | | Crystal structure of the Thermoplasma acidophilum Pelota protein | | Descriptor: | Cell division protein pelota related protein | | Authors: | Lee, H.H, Kim, Y.S, Kim, K.H, Heo, I.H, Kim, S.K, Kim, O, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into dom34, a key component of no-go mRNA decay
Mol.Cell, 27, 2007
|
|
1C2A
 
 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
6VJC
 
 | | LmFPPS mutant T164Y in complex with 476A, IPP & Ca | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Aripirala, S, Gabelli, S.B. | | Deposit date: | 2020-01-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6W7I
 
 | | LmFPPS mutant T164W in complex with 476A, IPP & Ca | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Aripirala, S, Gabelli, S.B. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6WW1
 
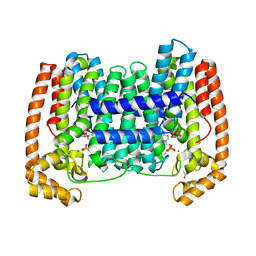 | | Crystal structure of the LmFPPS mutant E97Y | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Gabelli, S.B. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
2GKI
 
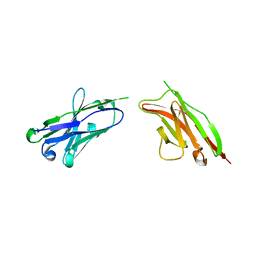 | | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity | | Descriptor: | nuclease | | Authors: | Kim, Y.R, Kim, J.S, Lee, S.H, Lee, W.R, Sohn, J.N, Chung, Y.C, Shim, H.K, Lee, S.C, Kwon, M.H, Kim, Y.S. | | Deposit date: | 2006-04-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Heavy and light chain variable single domains of an anti-DNA binding antibody hydrolyze both double- and single-stranded DNAs without sequence specificity.
J.Biol.Chem., 281, 2006
|
|
1DBG
 
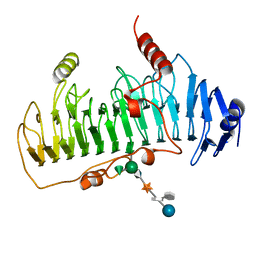 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1HMU
 
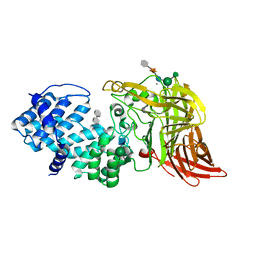 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HMW
 
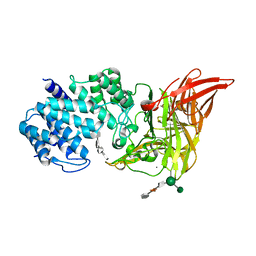 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HM3
 
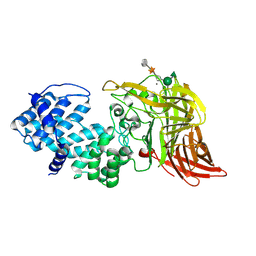 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1HM2
 
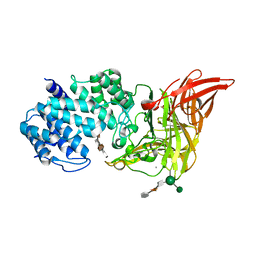 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1XNH
 
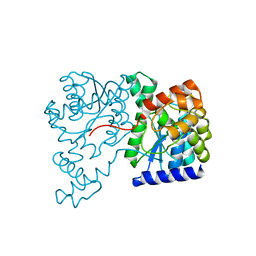 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1XNG
 
 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
3PGQ
 
 | | Crystal Structure of the Carboxyltransferase Domain of S. cerevisiae Acetyl CoA Carboxylase in Complex with Pinoxaden | | Descriptor: | 8-(2-ethenyl-6-ethyl-4-methylphenyl)tetrahydro-7H-pyrazolo[1,2-d][1,4,5]oxadiazepine-7,9(8H)-dione, Acetyl-CoA carboxylase | | Authors: | Tong, L, Yu, L.P.C, Kim, Y.S. | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins
Cell(Cambridge,Mass.), 186, 2023
|
|
4XSJ
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
