3ATT
 
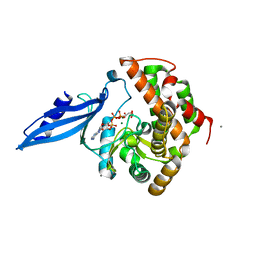 | | Crystal structure of Rv3168 with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
3ATS
 
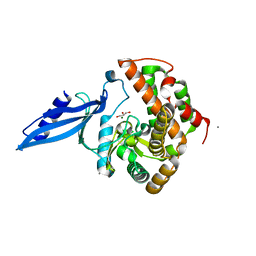 | | Crystal structure of Rv3168 | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
2WNW
 
 | | The crystal structure of SrfJ from salmonella typhimurium | | Descriptor: | ACTIVATED BY TRANSCRIPTION FACTOR SSRB, GLYCEROL, PHOSPHATE ION | | Authors: | Kim, Y.-G, Kim, J.-H, Kim, K.-J. | | Deposit date: | 2009-07-20 | | Release date: | 2010-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Salmonella Enterica Serovar Typhimurium Virulence Factor Srfj, a Glycoside Hydrolase Family Enzyme.
J.Bacteriol., 191, 2009
|
|
4N5L
 
 | |
4N5M
 
 | |
4N5N
 
 | |
5JJH
 
 | |
3AY3
 
 | |
2W8R
 
 | |
2W8Q
 
 | |
2W8N
 
 | |
2W8P
 
 | | The crystal structure of human C340A SSADH | | Descriptor: | GLYCEROL, SUCCINIC SEMIALDEHYDE DEHYDROGENASE MITOCHONDRIAL, SULFATE ION | | Authors: | Kim, Y.-G, Kim, K.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-09 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-Switch Modulation of Human Ssadh by Dynamic Catalytic Loop.
Embo J., 28, 2009
|
|
2W8O
 
 | |
7FBG
 
 | |
5GJN
 
 | |
5GJM
 
 | |
5GJP
 
 | |
5HXX
 
 | | Crystal structure of AspAT from Corynebacterium glutamicum | | Descriptor: | 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, ... | | Authors: | Son, H.-F, Kim, K.-J. | | Deposit date: | 2016-01-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into a novel class of aspartate aminotransferase from Corynebacterium glutamicum
To be published
|
|
5IJZ
 
 | |
7YA4
 
 | |
7YA3
 
 | |
7C18
 
 | |
7DIB
 
 | |
8HT2
 
 | |
8HT4
 
 | |
