3RO5
 
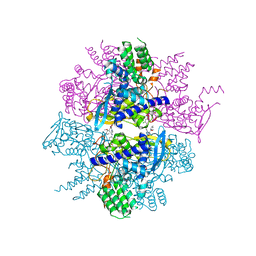 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1Z1W
 
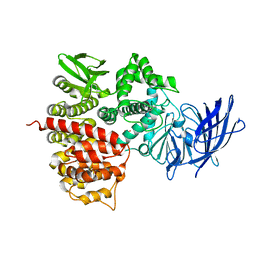 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1Z5H
 
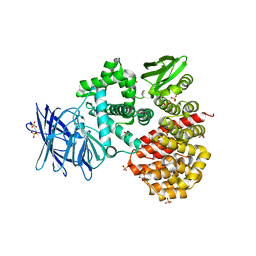 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1N5W
 
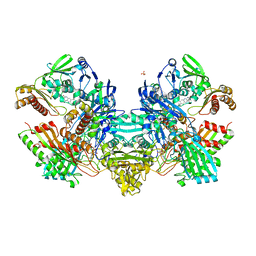 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Oxidized form | | Descriptor: | CU(I)-S-MO(VI)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-07 | | Release date: | 2002-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N63
 
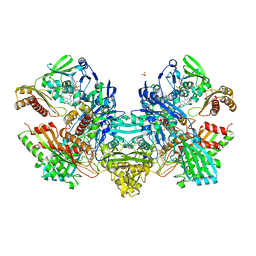 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Carbon monoxide reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N61
 
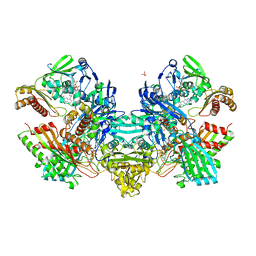 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Dithionite reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N62
 
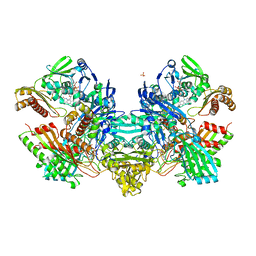 | | Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state | | Descriptor: | CU(I)-S-MO(IV)(=O)O-NBIC CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N60
 
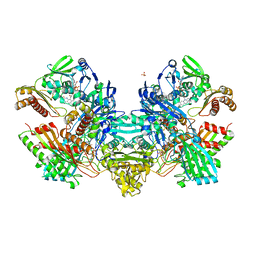 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Cyanide-inactivated Form | | Descriptor: | Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, Carbon monoxide dehydrogenase small chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ORW
 
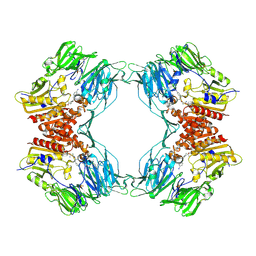 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P8J
 
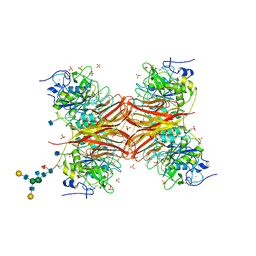 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
1ORV
 
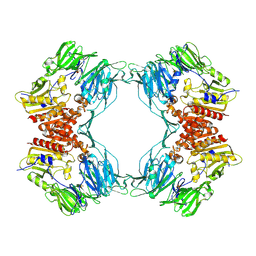 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SEZ
 
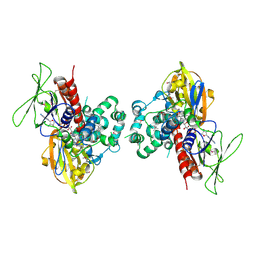 | | Crystal Structure of Protoporphyrinogen IX Oxidase | | Descriptor: | 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-BROMO-3-(5'-CARBOXY-4'-CHLORO-2'-FLUOROPHENYL)-1-METHYL-5-TRIFLUOROMETHYL-PYRAZOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Koch, M, Breithaupt, C, Kiefersauer, R, Freigang, J, Huber, R, Messerschmidt, A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
Embo J., 23, 2004
|
|
4DYS
 
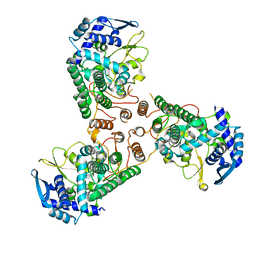 | | Crystal Structure of Apo Swine Flu Influenza Nucleoprotein | | Descriptor: | Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
4DYA
 
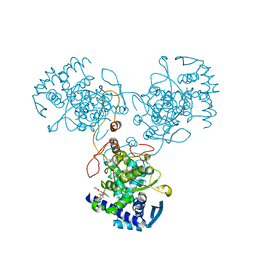 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885986 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]furan-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | To be determined
To be Published
|
|
4DYP
 
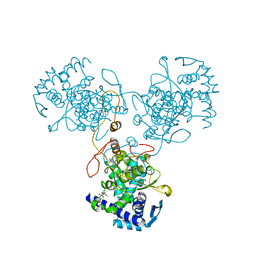 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-831780 Ligand Bound | | Descriptor: | Nucleocapsid protein, [4-(5-bromanyl-3-methyl-pyridin-2-yl)piperazin-1-yl]-[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | To be determined
To be Published
|
|
4DYT
 
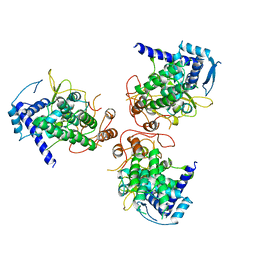 | | Crystal Structure of WSN/A Influenza Nucleoprotein with Three Mutations (E53D, Y289H, Y313V) | | Descriptor: | Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | To be determined
To be Published
|
|
4DYB
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-883559 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]thiophene-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
4DYN
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885838 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]pyridine-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | To be determined
To be Published
|
|
7ZXS
 
 | | Crystal structure of DPP9 in complex with a 4-oxo-b-lactam based inhibitor, A295 | | Descriptor: | 1,2-ETHANEDIOL, 2-ethyl-2-methanoyl-~{N}-[3-[[4-(quinolin-8-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7AYR
 
 | |
7AYQ
 
 | |
7OR4
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, B142 | | Descriptor: | Dipeptidyl peptidase 8, methyl 3-[4-[(4-bromophenyl)methyl]piperazin-1-yl]carbonyl-5-[(2-ethyl-2-methanoyl-butanoyl)amino]benzoate, trimethylamine oxide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OZ7
 
 | | Crystal structure of DPP8 in complex with a 4-oxo-b-lactam based inhibitor, L84 | | Descriptor: | Dipeptidyl peptidase 8, trimethylamine oxide, ~{N}-[3-[[4-[(4-bromophenyl)methyl]piperazin-1-yl]methyl]phenyl]-2-ethyl-2-methanoyl-butanamide | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-06-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1FQ3
 
 | | CRYSTAL STRUCTURE OF HUMAN GRANZYME B | | Descriptor: | GRANZYME B, SULFATE ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Estebanez-Perpina, E, Fuentes-Prior, P, Belorgey, D, Rubin, H, Bode, W. | | Deposit date: | 2000-09-03 | | Release date: | 2001-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the caspase activator human granzyme B, a proteinase highly specific for an Asp-P1 residue.
Biol.Chem., 381, 2000
|
|
6TRW
 
 | |
